
Epichloe festucae virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
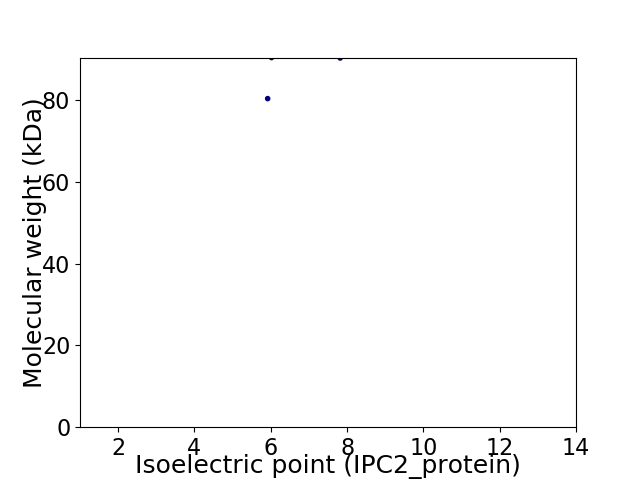
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1M2S1|Q1M2S1_9VIRU Putative coat protein OS=Epichloe festucae virus 1 OX=382962 GN=CP PE=4 SV=1
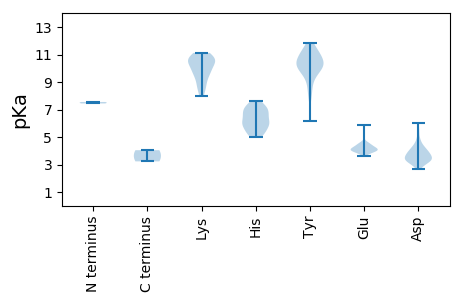
MM1 pKa = 7.44MSTAQTQNFQVRR13 pKa = 11.84HH14 pKa = 5.85LAPLTGAVADD24 pKa = 4.11PVGGLLQRR32 pKa = 11.84GKK34 pKa = 10.56DD35 pKa = 3.51FRR37 pKa = 11.84RR38 pKa = 11.84YY39 pKa = 9.5RR40 pKa = 11.84SGLSSEE46 pKa = 4.03IPGPGRR52 pKa = 11.84TFVEE56 pKa = 4.27YY57 pKa = 10.45RR58 pKa = 11.84SIFYY62 pKa = 10.41EE63 pKa = 4.06VGRR66 pKa = 11.84HH67 pKa = 5.01YY68 pKa = 11.66GNISDD73 pKa = 5.38ALATPTPAAVRR84 pKa = 11.84IDD86 pKa = 3.62CSHH89 pKa = 7.65TINEE93 pKa = 4.08TQAANFEE100 pKa = 4.33GLARR104 pKa = 11.84QYY106 pKa = 11.73SNFASQWEE114 pKa = 4.18KK115 pKa = 10.7MDD117 pKa = 3.44LCGIAEE123 pKa = 4.08RR124 pKa = 11.84LARR127 pKa = 11.84AIATQSVFGGVTTTHH142 pKa = 5.66MRR144 pKa = 11.84AGQPLRR150 pKa = 11.84VVALGTLDD158 pKa = 3.82SPQTASTSSVFIPRR172 pKa = 11.84TVDD175 pKa = 3.29SVGSDD180 pKa = 2.89HH181 pKa = 7.03VFAVLCAAANGEE193 pKa = 4.31GASVSTDD200 pKa = 3.2VLRR203 pKa = 11.84LDD205 pKa = 3.95VSNNAPLVPAVAGAPLATACVEE227 pKa = 4.0ALRR230 pKa = 11.84VLGANMEE237 pKa = 4.12EE238 pKa = 4.62SGAGDD243 pKa = 4.1LFAYY247 pKa = 10.12AVTRR251 pKa = 11.84GIHH254 pKa = 5.2RR255 pKa = 11.84VVTVVSHH262 pKa = 5.84TDD264 pKa = 2.73EE265 pKa = 5.1GGILRR270 pKa = 11.84DD271 pKa = 4.06LLRR274 pKa = 11.84HH275 pKa = 5.8DD276 pKa = 4.73AFRR279 pKa = 11.84VPYY282 pKa = 10.31GGINASLRR290 pKa = 11.84HH291 pKa = 5.31YY292 pKa = 9.18PALPSAAGSHH302 pKa = 6.11EE303 pKa = 4.25SSIAAWVDD311 pKa = 4.47GIALKK316 pKa = 9.22TAAVAAHH323 pKa = 7.18CDD325 pKa = 3.59PLVPGVGGMYY335 pKa = 7.99PTVLVASKK343 pKa = 11.04GSISPPGTPEE353 pKa = 4.35GDD355 pKa = 3.47VSPDD359 pKa = 3.35DD360 pKa = 3.81ARR362 pKa = 11.84SLGRR366 pKa = 11.84QLSADD371 pKa = 3.07IGRR374 pKa = 11.84FAPNYY379 pKa = 9.59IRR381 pKa = 11.84GLSALFGLQGYY392 pKa = 6.15TAIAEE397 pKa = 4.15RR398 pKa = 11.84HH399 pKa = 5.06LCATAGHH406 pKa = 5.72YY407 pKa = 10.66LEE409 pKa = 5.6EE410 pKa = 4.41NSATNRR416 pKa = 11.84HH417 pKa = 5.95LLHH420 pKa = 6.18KK421 pKa = 9.49TIAPYY426 pKa = 10.09FWVEE430 pKa = 3.89PTSLIPRR437 pKa = 11.84DD438 pKa = 3.77FLGTSAEE445 pKa = 4.23SEE447 pKa = 4.04NYY449 pKa = 8.32AAKK452 pKa = 10.48VSPGCVAHH460 pKa = 6.93EE461 pKa = 3.8PLFEE465 pKa = 4.31KK466 pKa = 10.36FAPLAQGTTMTCATAGFKK484 pKa = 9.69MRR486 pKa = 11.84TARR489 pKa = 11.84TSGYY493 pKa = 9.99VCSQAAAPASLAMIQLKK510 pKa = 9.86QFDD513 pKa = 4.14TQSVVLAGDD522 pKa = 3.36QGPISGDD529 pKa = 3.15VVTKK533 pKa = 10.43HH534 pKa = 5.97RR535 pKa = 11.84ASSPLSSYY543 pKa = 10.78LWVRR547 pKa = 11.84GQSALPAPAEE557 pKa = 4.25FINTQGCYY565 pKa = 8.63GAKK568 pKa = 9.7IKK570 pKa = 10.94LVDD573 pKa = 3.86WTDD576 pKa = 3.21DD577 pKa = 3.38WDD579 pKa = 4.29PEE581 pKa = 4.47STDD584 pKa = 3.45VPRR587 pKa = 11.84PDD589 pKa = 3.24QFATGVVAWRR599 pKa = 11.84VSLPTGLPTGPSNAADD615 pKa = 3.79RR616 pKa = 11.84QARR619 pKa = 11.84RR620 pKa = 11.84ARR622 pKa = 11.84SRR624 pKa = 11.84GAIALAQALNRR635 pKa = 11.84ARR637 pKa = 11.84AFGEE641 pKa = 4.47GASPTMEE648 pKa = 4.03VSDD651 pKa = 4.24VPPDD655 pKa = 3.93LGLADD660 pKa = 4.84LGHH663 pKa = 6.59YY664 pKa = 10.12DD665 pKa = 3.11HH666 pKa = 7.23TGFRR670 pKa = 11.84EE671 pKa = 3.93HH672 pKa = 7.42RR673 pKa = 11.84SDD675 pKa = 3.39PGKK678 pKa = 10.57AQEE681 pKa = 4.62TGDD684 pKa = 4.24GNPSPTPTLLRR695 pKa = 11.84GAAMPPTEE703 pKa = 3.78QHH705 pKa = 6.31KK706 pKa = 9.45PQGGPRR712 pKa = 11.84LPAPPPTMGRR722 pKa = 11.84AAGPVGPPSTPAPPPPAPVIAPPTVSDD749 pKa = 4.28PPPMPEE755 pKa = 3.73AATGAEE761 pKa = 4.39PVPQAA766 pKa = 4.04
MM1 pKa = 7.44MSTAQTQNFQVRR13 pKa = 11.84HH14 pKa = 5.85LAPLTGAVADD24 pKa = 4.11PVGGLLQRR32 pKa = 11.84GKK34 pKa = 10.56DD35 pKa = 3.51FRR37 pKa = 11.84RR38 pKa = 11.84YY39 pKa = 9.5RR40 pKa = 11.84SGLSSEE46 pKa = 4.03IPGPGRR52 pKa = 11.84TFVEE56 pKa = 4.27YY57 pKa = 10.45RR58 pKa = 11.84SIFYY62 pKa = 10.41EE63 pKa = 4.06VGRR66 pKa = 11.84HH67 pKa = 5.01YY68 pKa = 11.66GNISDD73 pKa = 5.38ALATPTPAAVRR84 pKa = 11.84IDD86 pKa = 3.62CSHH89 pKa = 7.65TINEE93 pKa = 4.08TQAANFEE100 pKa = 4.33GLARR104 pKa = 11.84QYY106 pKa = 11.73SNFASQWEE114 pKa = 4.18KK115 pKa = 10.7MDD117 pKa = 3.44LCGIAEE123 pKa = 4.08RR124 pKa = 11.84LARR127 pKa = 11.84AIATQSVFGGVTTTHH142 pKa = 5.66MRR144 pKa = 11.84AGQPLRR150 pKa = 11.84VVALGTLDD158 pKa = 3.82SPQTASTSSVFIPRR172 pKa = 11.84TVDD175 pKa = 3.29SVGSDD180 pKa = 2.89HH181 pKa = 7.03VFAVLCAAANGEE193 pKa = 4.31GASVSTDD200 pKa = 3.2VLRR203 pKa = 11.84LDD205 pKa = 3.95VSNNAPLVPAVAGAPLATACVEE227 pKa = 4.0ALRR230 pKa = 11.84VLGANMEE237 pKa = 4.12EE238 pKa = 4.62SGAGDD243 pKa = 4.1LFAYY247 pKa = 10.12AVTRR251 pKa = 11.84GIHH254 pKa = 5.2RR255 pKa = 11.84VVTVVSHH262 pKa = 5.84TDD264 pKa = 2.73EE265 pKa = 5.1GGILRR270 pKa = 11.84DD271 pKa = 4.06LLRR274 pKa = 11.84HH275 pKa = 5.8DD276 pKa = 4.73AFRR279 pKa = 11.84VPYY282 pKa = 10.31GGINASLRR290 pKa = 11.84HH291 pKa = 5.31YY292 pKa = 9.18PALPSAAGSHH302 pKa = 6.11EE303 pKa = 4.25SSIAAWVDD311 pKa = 4.47GIALKK316 pKa = 9.22TAAVAAHH323 pKa = 7.18CDD325 pKa = 3.59PLVPGVGGMYY335 pKa = 7.99PTVLVASKK343 pKa = 11.04GSISPPGTPEE353 pKa = 4.35GDD355 pKa = 3.47VSPDD359 pKa = 3.35DD360 pKa = 3.81ARR362 pKa = 11.84SLGRR366 pKa = 11.84QLSADD371 pKa = 3.07IGRR374 pKa = 11.84FAPNYY379 pKa = 9.59IRR381 pKa = 11.84GLSALFGLQGYY392 pKa = 6.15TAIAEE397 pKa = 4.15RR398 pKa = 11.84HH399 pKa = 5.06LCATAGHH406 pKa = 5.72YY407 pKa = 10.66LEE409 pKa = 5.6EE410 pKa = 4.41NSATNRR416 pKa = 11.84HH417 pKa = 5.95LLHH420 pKa = 6.18KK421 pKa = 9.49TIAPYY426 pKa = 10.09FWVEE430 pKa = 3.89PTSLIPRR437 pKa = 11.84DD438 pKa = 3.77FLGTSAEE445 pKa = 4.23SEE447 pKa = 4.04NYY449 pKa = 8.32AAKK452 pKa = 10.48VSPGCVAHH460 pKa = 6.93EE461 pKa = 3.8PLFEE465 pKa = 4.31KK466 pKa = 10.36FAPLAQGTTMTCATAGFKK484 pKa = 9.69MRR486 pKa = 11.84TARR489 pKa = 11.84TSGYY493 pKa = 9.99VCSQAAAPASLAMIQLKK510 pKa = 9.86QFDD513 pKa = 4.14TQSVVLAGDD522 pKa = 3.36QGPISGDD529 pKa = 3.15VVTKK533 pKa = 10.43HH534 pKa = 5.97RR535 pKa = 11.84ASSPLSSYY543 pKa = 10.78LWVRR547 pKa = 11.84GQSALPAPAEE557 pKa = 4.25FINTQGCYY565 pKa = 8.63GAKK568 pKa = 9.7IKK570 pKa = 10.94LVDD573 pKa = 3.86WTDD576 pKa = 3.21DD577 pKa = 3.38WDD579 pKa = 4.29PEE581 pKa = 4.47STDD584 pKa = 3.45VPRR587 pKa = 11.84PDD589 pKa = 3.24QFATGVVAWRR599 pKa = 11.84VSLPTGLPTGPSNAADD615 pKa = 3.79RR616 pKa = 11.84QARR619 pKa = 11.84RR620 pKa = 11.84ARR622 pKa = 11.84SRR624 pKa = 11.84GAIALAQALNRR635 pKa = 11.84ARR637 pKa = 11.84AFGEE641 pKa = 4.47GASPTMEE648 pKa = 4.03VSDD651 pKa = 4.24VPPDD655 pKa = 3.93LGLADD660 pKa = 4.84LGHH663 pKa = 6.59YY664 pKa = 10.12DD665 pKa = 3.11HH666 pKa = 7.23TGFRR670 pKa = 11.84EE671 pKa = 3.93HH672 pKa = 7.42RR673 pKa = 11.84SDD675 pKa = 3.39PGKK678 pKa = 10.57AQEE681 pKa = 4.62TGDD684 pKa = 4.24GNPSPTPTLLRR695 pKa = 11.84GAAMPPTEE703 pKa = 3.78QHH705 pKa = 6.31KK706 pKa = 9.45PQGGPRR712 pKa = 11.84LPAPPPTMGRR722 pKa = 11.84AAGPVGPPSTPAPPPPAPVIAPPTVSDD749 pKa = 4.28PPPMPEE755 pKa = 3.73AATGAEE761 pKa = 4.39PVPQAA766 pKa = 4.04
Molecular weight: 80.3 kDa
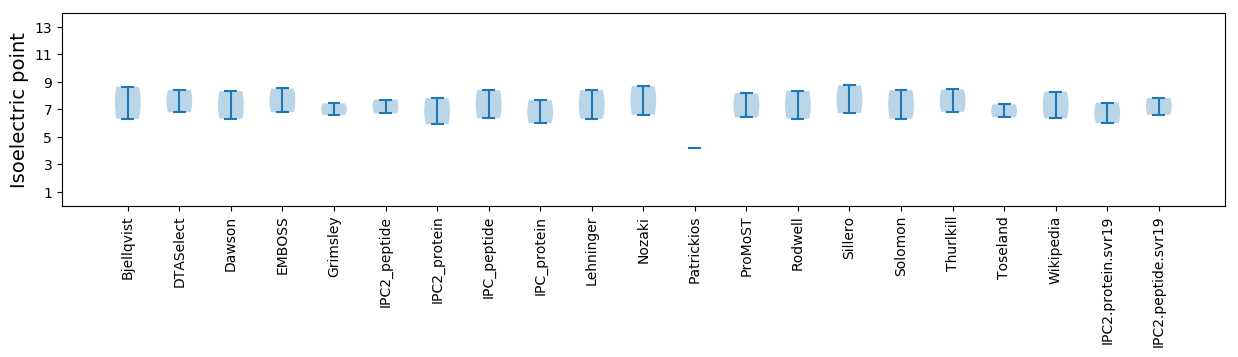
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q1M2S1|Q1M2S1_9VIRU Putative coat protein OS=Epichloe festucae virus 1 OX=382962 GN=CP PE=4 SV=1
MM1 pKa = 7.55IEE3 pKa = 3.94STVADD8 pKa = 4.17RR9 pKa = 11.84ANAAGSVGRR18 pKa = 11.84YY19 pKa = 9.4LEE21 pKa = 4.35ALLPSEE27 pKa = 4.2YY28 pKa = 10.09TRR30 pKa = 11.84VVSTLPFDD38 pKa = 3.8HH39 pKa = 6.37QVSFVYY45 pKa = 10.27RR46 pKa = 11.84PSWRR50 pKa = 11.84HH51 pKa = 5.14HH52 pKa = 6.54KK53 pKa = 8.77PTEE56 pKa = 4.0LQRR59 pKa = 11.84VAAAFLLPRR68 pKa = 11.84VPVQVSYY75 pKa = 11.13SAGSILTLLNATLPNLPPRR94 pKa = 11.84HH95 pKa = 6.11TYY97 pKa = 8.72HH98 pKa = 7.15AGWSCDD104 pKa = 2.97SDD106 pKa = 3.55ATRR109 pKa = 11.84KK110 pKa = 9.78AFPLKK115 pKa = 10.69ANPAASNKK123 pKa = 8.79VNVYY127 pKa = 10.31LSEE130 pKa = 3.87VCYY133 pKa = 10.4DD134 pKa = 3.9LRR136 pKa = 11.84RR137 pKa = 11.84TNPGEE142 pKa = 4.0AMAAEE147 pKa = 4.57AALQRR152 pKa = 11.84VRR154 pKa = 11.84SSGSVYY160 pKa = 10.56NDD162 pKa = 2.77QATAVVLYY170 pKa = 7.78GFCLARR176 pKa = 11.84NGLPGAYY183 pKa = 9.84DD184 pKa = 3.06IALALLLNPDD194 pKa = 3.73YY195 pKa = 11.1AKK197 pKa = 10.96SLTGVLKK204 pKa = 10.09ATGGNATAVGSLLVEE219 pKa = 4.5ANTLLGRR226 pKa = 11.84DD227 pKa = 3.85VAPVDD232 pKa = 3.83LGKK235 pKa = 10.38EE236 pKa = 4.14FNMRR240 pKa = 11.84TQLRR244 pKa = 11.84LARR247 pKa = 11.84EE248 pKa = 4.03NMAEE252 pKa = 4.09YY253 pKa = 9.98PPDD256 pKa = 3.5VLRR259 pKa = 11.84ATVRR263 pKa = 11.84RR264 pKa = 11.84VLLDD268 pKa = 3.2EE269 pKa = 4.07LTRR272 pKa = 11.84RR273 pKa = 11.84AGSYY277 pKa = 10.01EE278 pKa = 4.08LDD280 pKa = 3.63YY281 pKa = 11.2PSLEE285 pKa = 4.25EE286 pKa = 4.1HH287 pKa = 6.8WDD289 pKa = 3.81SRR291 pKa = 11.84WAWAVNGAQGGAITDD306 pKa = 3.9KK307 pKa = 10.91GLEE310 pKa = 4.18SAPRR314 pKa = 11.84PPGATRR320 pKa = 11.84EE321 pKa = 4.07YY322 pKa = 9.88RR323 pKa = 11.84RR324 pKa = 11.84SWLEE328 pKa = 3.65RR329 pKa = 11.84VEE331 pKa = 4.12EE332 pKa = 4.21DD333 pKa = 5.49PRR335 pKa = 11.84PQWDD339 pKa = 3.5GTTEE343 pKa = 3.89VSASSKK349 pKa = 10.62LEE351 pKa = 3.94HH352 pKa = 6.53GKK354 pKa = 7.98TRR356 pKa = 11.84AIFACDD362 pKa = 3.2TVNYY366 pKa = 10.14LAFEE370 pKa = 4.5HH371 pKa = 6.76LMSTVEE377 pKa = 4.06ANWRR381 pKa = 11.84GVRR384 pKa = 11.84VVLNPGKK391 pKa = 10.15GGNMGMAMRR400 pKa = 11.84VQAARR405 pKa = 11.84RR406 pKa = 11.84RR407 pKa = 11.84CGVSLMLDD415 pKa = 3.2YY416 pKa = 11.57DD417 pKa = 4.5DD418 pKa = 5.99FNSHH422 pKa = 5.4HH423 pKa = 7.22TISAMQIVVEE433 pKa = 4.56EE434 pKa = 4.46VCSITGYY441 pKa = 11.11DD442 pKa = 3.48PDD444 pKa = 3.59LAARR448 pKa = 11.84LVASLDD454 pKa = 3.42KK455 pKa = 11.09QYY457 pKa = 9.7ITLAGRR463 pKa = 11.84RR464 pKa = 11.84KK465 pKa = 10.21LSVGTLMSGHH475 pKa = 6.91RR476 pKa = 11.84CTTFFNSVLNAVYY489 pKa = 10.43VRR491 pKa = 11.84LEE493 pKa = 4.13LGEE496 pKa = 4.22PFYY499 pKa = 11.48SEE501 pKa = 4.26TVSLHH506 pKa = 5.96VGDD509 pKa = 4.41DD510 pKa = 3.37VYY512 pKa = 10.99MGVRR516 pKa = 11.84DD517 pKa = 4.23YY518 pKa = 11.86VSAAYY523 pKa = 9.86VIQRR527 pKa = 11.84LADD530 pKa = 3.29SPLRR534 pKa = 11.84MNALKK539 pKa = 10.59QSVGHH544 pKa = 6.11VSTEE548 pKa = 3.76FLRR551 pKa = 11.84IATGARR557 pKa = 11.84DD558 pKa = 3.57CYY560 pKa = 10.89GYY562 pKa = 10.97LSRR565 pKa = 11.84AVASIVSGNWVNDD578 pKa = 3.31TALAPFEE585 pKa = 4.52GLAAMVASARR595 pKa = 11.84SLANRR600 pKa = 11.84GKK602 pKa = 9.19TNLAPLLLVSSVLRR616 pKa = 11.84IAKK619 pKa = 9.69LGKK622 pKa = 8.53RR623 pKa = 11.84HH624 pKa = 5.43YY625 pKa = 10.56GKK627 pKa = 10.62VSSLLDD633 pKa = 3.32GTVALGNGPQYY644 pKa = 11.27SCGAHH649 pKa = 5.51MRR651 pKa = 11.84WCNPVVVRR659 pKa = 11.84DD660 pKa = 4.03TPDD663 pKa = 2.67PWGYY667 pKa = 9.12TEE669 pKa = 5.9LPLEE673 pKa = 4.36SVTAYY678 pKa = 10.69LSRR681 pKa = 11.84AASPLEE687 pKa = 3.91WEE689 pKa = 4.6VLGEE693 pKa = 4.01AGISVTEE700 pKa = 3.89QMAAASYY707 pKa = 9.53RR708 pKa = 11.84KK709 pKa = 8.18TYY711 pKa = 11.13ASLFWRR717 pKa = 11.84GEE719 pKa = 3.94RR720 pKa = 11.84LTLGPTLSKK729 pKa = 11.12APIGSASAEE738 pKa = 3.96DD739 pKa = 3.79LARR742 pKa = 11.84ATKK745 pKa = 9.9PHH747 pKa = 6.69GLLAQYY753 pKa = 10.24PLLLLARR760 pKa = 11.84DD761 pKa = 4.98RR762 pKa = 11.84IPEE765 pKa = 4.03TLVRR769 pKa = 11.84VALGKK774 pKa = 10.64AGGNPHH780 pKa = 6.59TSHH783 pKa = 7.5LAYY786 pKa = 10.29DD787 pKa = 3.29AWGEE791 pKa = 4.01YY792 pKa = 10.57DD793 pKa = 4.65HH794 pKa = 7.06GCQIQTVLGYY804 pKa = 11.03ADD806 pKa = 3.57AAAFGKK812 pKa = 8.69RR813 pKa = 11.84TSADD817 pKa = 3.13VLTSRR822 pKa = 11.84TLMYY826 pKa = 10.73VV827 pKa = 3.23
MM1 pKa = 7.55IEE3 pKa = 3.94STVADD8 pKa = 4.17RR9 pKa = 11.84ANAAGSVGRR18 pKa = 11.84YY19 pKa = 9.4LEE21 pKa = 4.35ALLPSEE27 pKa = 4.2YY28 pKa = 10.09TRR30 pKa = 11.84VVSTLPFDD38 pKa = 3.8HH39 pKa = 6.37QVSFVYY45 pKa = 10.27RR46 pKa = 11.84PSWRR50 pKa = 11.84HH51 pKa = 5.14HH52 pKa = 6.54KK53 pKa = 8.77PTEE56 pKa = 4.0LQRR59 pKa = 11.84VAAAFLLPRR68 pKa = 11.84VPVQVSYY75 pKa = 11.13SAGSILTLLNATLPNLPPRR94 pKa = 11.84HH95 pKa = 6.11TYY97 pKa = 8.72HH98 pKa = 7.15AGWSCDD104 pKa = 2.97SDD106 pKa = 3.55ATRR109 pKa = 11.84KK110 pKa = 9.78AFPLKK115 pKa = 10.69ANPAASNKK123 pKa = 8.79VNVYY127 pKa = 10.31LSEE130 pKa = 3.87VCYY133 pKa = 10.4DD134 pKa = 3.9LRR136 pKa = 11.84RR137 pKa = 11.84TNPGEE142 pKa = 4.0AMAAEE147 pKa = 4.57AALQRR152 pKa = 11.84VRR154 pKa = 11.84SSGSVYY160 pKa = 10.56NDD162 pKa = 2.77QATAVVLYY170 pKa = 7.78GFCLARR176 pKa = 11.84NGLPGAYY183 pKa = 9.84DD184 pKa = 3.06IALALLLNPDD194 pKa = 3.73YY195 pKa = 11.1AKK197 pKa = 10.96SLTGVLKK204 pKa = 10.09ATGGNATAVGSLLVEE219 pKa = 4.5ANTLLGRR226 pKa = 11.84DD227 pKa = 3.85VAPVDD232 pKa = 3.83LGKK235 pKa = 10.38EE236 pKa = 4.14FNMRR240 pKa = 11.84TQLRR244 pKa = 11.84LARR247 pKa = 11.84EE248 pKa = 4.03NMAEE252 pKa = 4.09YY253 pKa = 9.98PPDD256 pKa = 3.5VLRR259 pKa = 11.84ATVRR263 pKa = 11.84RR264 pKa = 11.84VLLDD268 pKa = 3.2EE269 pKa = 4.07LTRR272 pKa = 11.84RR273 pKa = 11.84AGSYY277 pKa = 10.01EE278 pKa = 4.08LDD280 pKa = 3.63YY281 pKa = 11.2PSLEE285 pKa = 4.25EE286 pKa = 4.1HH287 pKa = 6.8WDD289 pKa = 3.81SRR291 pKa = 11.84WAWAVNGAQGGAITDD306 pKa = 3.9KK307 pKa = 10.91GLEE310 pKa = 4.18SAPRR314 pKa = 11.84PPGATRR320 pKa = 11.84EE321 pKa = 4.07YY322 pKa = 9.88RR323 pKa = 11.84RR324 pKa = 11.84SWLEE328 pKa = 3.65RR329 pKa = 11.84VEE331 pKa = 4.12EE332 pKa = 4.21DD333 pKa = 5.49PRR335 pKa = 11.84PQWDD339 pKa = 3.5GTTEE343 pKa = 3.89VSASSKK349 pKa = 10.62LEE351 pKa = 3.94HH352 pKa = 6.53GKK354 pKa = 7.98TRR356 pKa = 11.84AIFACDD362 pKa = 3.2TVNYY366 pKa = 10.14LAFEE370 pKa = 4.5HH371 pKa = 6.76LMSTVEE377 pKa = 4.06ANWRR381 pKa = 11.84GVRR384 pKa = 11.84VVLNPGKK391 pKa = 10.15GGNMGMAMRR400 pKa = 11.84VQAARR405 pKa = 11.84RR406 pKa = 11.84RR407 pKa = 11.84CGVSLMLDD415 pKa = 3.2YY416 pKa = 11.57DD417 pKa = 4.5DD418 pKa = 5.99FNSHH422 pKa = 5.4HH423 pKa = 7.22TISAMQIVVEE433 pKa = 4.56EE434 pKa = 4.46VCSITGYY441 pKa = 11.11DD442 pKa = 3.48PDD444 pKa = 3.59LAARR448 pKa = 11.84LVASLDD454 pKa = 3.42KK455 pKa = 11.09QYY457 pKa = 9.7ITLAGRR463 pKa = 11.84RR464 pKa = 11.84KK465 pKa = 10.21LSVGTLMSGHH475 pKa = 6.91RR476 pKa = 11.84CTTFFNSVLNAVYY489 pKa = 10.43VRR491 pKa = 11.84LEE493 pKa = 4.13LGEE496 pKa = 4.22PFYY499 pKa = 11.48SEE501 pKa = 4.26TVSLHH506 pKa = 5.96VGDD509 pKa = 4.41DD510 pKa = 3.37VYY512 pKa = 10.99MGVRR516 pKa = 11.84DD517 pKa = 4.23YY518 pKa = 11.86VSAAYY523 pKa = 9.86VIQRR527 pKa = 11.84LADD530 pKa = 3.29SPLRR534 pKa = 11.84MNALKK539 pKa = 10.59QSVGHH544 pKa = 6.11VSTEE548 pKa = 3.76FLRR551 pKa = 11.84IATGARR557 pKa = 11.84DD558 pKa = 3.57CYY560 pKa = 10.89GYY562 pKa = 10.97LSRR565 pKa = 11.84AVASIVSGNWVNDD578 pKa = 3.31TALAPFEE585 pKa = 4.52GLAAMVASARR595 pKa = 11.84SLANRR600 pKa = 11.84GKK602 pKa = 9.19TNLAPLLLVSSVLRR616 pKa = 11.84IAKK619 pKa = 9.69LGKK622 pKa = 8.53RR623 pKa = 11.84HH624 pKa = 5.43YY625 pKa = 10.56GKK627 pKa = 10.62VSSLLDD633 pKa = 3.32GTVALGNGPQYY644 pKa = 11.27SCGAHH649 pKa = 5.51MRR651 pKa = 11.84WCNPVVVRR659 pKa = 11.84DD660 pKa = 4.03TPDD663 pKa = 2.67PWGYY667 pKa = 9.12TEE669 pKa = 5.9LPLEE673 pKa = 4.36SVTAYY678 pKa = 10.69LSRR681 pKa = 11.84AASPLEE687 pKa = 3.91WEE689 pKa = 4.6VLGEE693 pKa = 4.01AGISVTEE700 pKa = 3.89QMAAASYY707 pKa = 9.53RR708 pKa = 11.84KK709 pKa = 8.18TYY711 pKa = 11.13ASLFWRR717 pKa = 11.84GEE719 pKa = 3.94RR720 pKa = 11.84LTLGPTLSKK729 pKa = 11.12APIGSASAEE738 pKa = 3.96DD739 pKa = 3.79LARR742 pKa = 11.84ATKK745 pKa = 9.9PHH747 pKa = 6.69GLLAQYY753 pKa = 10.24PLLLLARR760 pKa = 11.84DD761 pKa = 4.98RR762 pKa = 11.84IPEE765 pKa = 4.03TLVRR769 pKa = 11.84VALGKK774 pKa = 10.64AGGNPHH780 pKa = 6.59TSHH783 pKa = 7.5LAYY786 pKa = 10.29DD787 pKa = 3.29AWGEE791 pKa = 4.01YY792 pKa = 10.57DD793 pKa = 4.65HH794 pKa = 7.06GCQIQTVLGYY804 pKa = 11.03ADD806 pKa = 3.57AAAFGKK812 pKa = 8.69RR813 pKa = 11.84TSADD817 pKa = 3.13VLTSRR822 pKa = 11.84TLMYY826 pKa = 10.73VV827 pKa = 3.23
Molecular weight: 90.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1593 |
766 |
827 |
796.5 |
85.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.869 ± 0.576 | 1.318 ± 0.009 |
4.834 ± 0.087 | 4.52 ± 0.325 |
2.448 ± 0.381 | 8.349 ± 0.632 |
2.511 ± 0.158 | 2.637 ± 0.431 |
2.323 ± 0.34 | 9.605 ± 1.395 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.883 ± 0.128 | 3.013 ± 0.455 |
7.094 ± 1.583 | 2.825 ± 0.481 |
7.031 ± 0.525 | 7.596 ± 0.016 |
6.529 ± 0.358 | 7.847 ± 0.458 |
1.318 ± 0.278 | 3.453 ± 0.757 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
