
Escherichia phage Lilleput
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; Bullavirinae; unclassified Bullavirinae
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
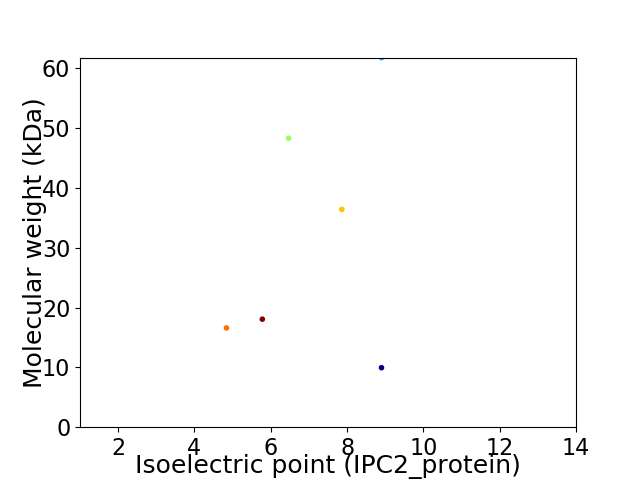
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
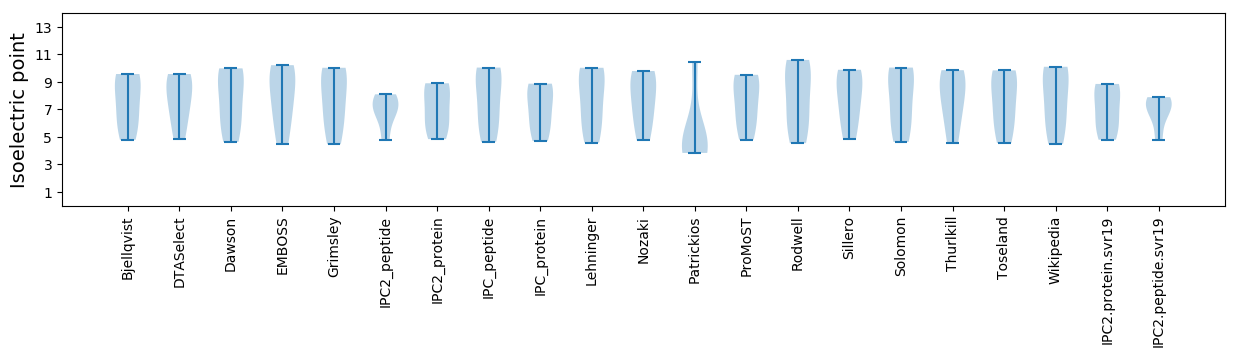
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6DZ07|A0A4D6DZ07_9VIRU H protein OS=Escherichia phage Lilleput OX=2562232 PE=4 SV=1
MM1 pKa = 7.53NSNEE5 pKa = 4.22SAVAFQTAIASIKK18 pKa = 9.2MIQASSVLDD27 pKa = 3.53LTEE30 pKa = 5.82DD31 pKa = 3.47DD32 pKa = 5.78FEE34 pKa = 4.82FLTGDD39 pKa = 3.9RR40 pKa = 11.84VWIATDD46 pKa = 3.49RR47 pKa = 11.84SRR49 pKa = 11.84ARR51 pKa = 11.84RR52 pKa = 11.84AVEE55 pKa = 3.52ACVYY59 pKa = 8.38GTLDD63 pKa = 3.27FVGYY67 pKa = 9.24PRR69 pKa = 11.84FPAPVEE75 pKa = 4.42FISAVIAYY83 pKa = 7.79YY84 pKa = 8.7VHH86 pKa = 6.96PVNIQTACLIMEE98 pKa = 4.57GAEE101 pKa = 3.88FTEE104 pKa = 4.67NIVNGVEE111 pKa = 4.44SPVKK115 pKa = 10.44ASEE118 pKa = 3.96LFAFVLRR125 pKa = 11.84VRR127 pKa = 11.84AGCKK131 pKa = 9.8DD132 pKa = 3.6VLEE135 pKa = 4.51GAEE138 pKa = 3.99DD139 pKa = 3.92NIRR142 pKa = 11.84AKK144 pKa = 10.71LRR146 pKa = 11.84AQGIMM151 pKa = 3.61
MM1 pKa = 7.53NSNEE5 pKa = 4.22SAVAFQTAIASIKK18 pKa = 9.2MIQASSVLDD27 pKa = 3.53LTEE30 pKa = 5.82DD31 pKa = 3.47DD32 pKa = 5.78FEE34 pKa = 4.82FLTGDD39 pKa = 3.9RR40 pKa = 11.84VWIATDD46 pKa = 3.49RR47 pKa = 11.84SRR49 pKa = 11.84ARR51 pKa = 11.84RR52 pKa = 11.84AVEE55 pKa = 3.52ACVYY59 pKa = 8.38GTLDD63 pKa = 3.27FVGYY67 pKa = 9.24PRR69 pKa = 11.84FPAPVEE75 pKa = 4.42FISAVIAYY83 pKa = 7.79YY84 pKa = 8.7VHH86 pKa = 6.96PVNIQTACLIMEE98 pKa = 4.57GAEE101 pKa = 3.88FTEE104 pKa = 4.67NIVNGVEE111 pKa = 4.44SPVKK115 pKa = 10.44ASEE118 pKa = 3.96LFAFVLRR125 pKa = 11.84VRR127 pKa = 11.84AGCKK131 pKa = 9.8DD132 pKa = 3.6VLEE135 pKa = 4.51GAEE138 pKa = 3.99DD139 pKa = 3.92NIRR142 pKa = 11.84AKK144 pKa = 10.71LRR146 pKa = 11.84AQGIMM151 pKa = 3.61
Molecular weight: 16.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6DYW2|A0A4D6DYW2_9VIRU External scaffolding protein D OS=Escherichia phage Lilleput OX=2562232 PE=3 SV=1
MM1 pKa = 7.44SVEE4 pKa = 4.3LIKK7 pKa = 10.7PLVEE11 pKa = 3.86AAGMSTLTQSPNLRR25 pKa = 11.84IWKK28 pKa = 8.93QVNTRR33 pKa = 11.84VKK35 pKa = 10.51LLEE38 pKa = 4.88EE39 pKa = 4.62ILAHH43 pKa = 5.1WTNGIRR49 pKa = 11.84RR50 pKa = 11.84CPEE53 pKa = 3.56TQDD56 pKa = 4.78FYY58 pKa = 11.45MNPNSQLATTIAYY71 pKa = 8.04RR72 pKa = 11.84AHH74 pKa = 6.19QKK76 pKa = 9.51GHH78 pKa = 5.39NPKK81 pKa = 10.28LIQYY85 pKa = 8.27PDD87 pKa = 3.65TFTLDD92 pKa = 3.71DD93 pKa = 4.6IIRR96 pKa = 11.84RR97 pKa = 11.84KK98 pKa = 9.8PIPQTAPNEE107 pKa = 3.94LRR109 pKa = 11.84LSDD112 pKa = 5.03EE113 pKa = 3.69IGEE116 pKa = 4.37DD117 pKa = 3.51YY118 pKa = 10.75RR119 pKa = 11.84LTVIAIADD127 pKa = 4.11EE128 pKa = 4.23LQEE131 pKa = 4.7CYY133 pKa = 10.51DD134 pKa = 3.64VLGQLDD140 pKa = 3.83INNTIDD146 pKa = 3.4HH147 pKa = 6.15TPKK150 pKa = 10.95GNAHH154 pKa = 6.5WSMLYY159 pKa = 10.22DD160 pKa = 4.05KK161 pKa = 10.32PVNTHH166 pKa = 5.13WSQLVSKK173 pKa = 10.58RR174 pKa = 11.84PLKK177 pKa = 10.43DD178 pKa = 2.77IRR180 pKa = 11.84ADD182 pKa = 3.47YY183 pKa = 10.94NYY185 pKa = 11.09AKK187 pKa = 10.74AKK189 pKa = 9.85GIKK192 pKa = 9.69DD193 pKa = 3.59EE194 pKa = 4.13CSKK197 pKa = 10.4MLEE200 pKa = 4.05EE201 pKa = 4.36STMKK205 pKa = 10.09SRR207 pKa = 11.84RR208 pKa = 11.84GFTVQRR214 pKa = 11.84LMNAMRR220 pKa = 11.84QAHH223 pKa = 5.52TDD225 pKa = 2.56GWYY228 pKa = 10.64VVFDD232 pKa = 4.23TLTLADD238 pKa = 4.35DD239 pKa = 5.61RR240 pKa = 11.84IQAFYY245 pKa = 11.18DD246 pKa = 3.46NPNALRR252 pKa = 11.84DD253 pKa = 3.67YY254 pKa = 10.67FRR256 pKa = 11.84DD257 pKa = 3.04IGRR260 pKa = 11.84MVLAAEE266 pKa = 4.55GRR268 pKa = 11.84SANDD272 pKa = 3.16SHH274 pKa = 7.55SDD276 pKa = 3.48CYY278 pKa = 11.08QYY280 pKa = 11.34FCVPEE285 pKa = 4.02YY286 pKa = 9.72GTQHH290 pKa = 5.68GRR292 pKa = 11.84LHH294 pKa = 5.96FHH296 pKa = 6.87AVHH299 pKa = 6.99LMRR302 pKa = 11.84TLPLGSRR309 pKa = 11.84DD310 pKa = 3.68PNFGKK315 pKa = 10.52LVRR318 pKa = 11.84TNRR321 pKa = 11.84QINSLQNTWPYY332 pKa = 9.75GYY334 pKa = 10.78SMPIAVRR341 pKa = 11.84YY342 pKa = 9.26SQDD345 pKa = 2.82AFSRR349 pKa = 11.84AGWLWPVDD357 pKa = 4.15SKK359 pKa = 11.92GEE361 pKa = 4.01PLKK364 pKa = 10.22ATSYY368 pKa = 9.01MAVGFYY374 pKa = 9.16VAKK377 pKa = 10.34YY378 pKa = 9.45VNKK381 pKa = 10.55KK382 pKa = 10.41SDD384 pKa = 2.79IDD386 pKa = 3.67MAAKK390 pKa = 10.6GLGNKK395 pKa = 7.49EE396 pKa = 3.68WNASLKK402 pKa = 9.91TKK404 pKa = 10.52ISLLPTKK411 pKa = 10.29LFRR414 pKa = 11.84IRR416 pKa = 11.84MSRR419 pKa = 11.84NFGMKK424 pKa = 8.53MLSMAHH430 pKa = 5.91LTAEE434 pKa = 4.48ALIQLTQLGYY444 pKa = 11.13DD445 pKa = 3.19STPFNNVLKK454 pKa = 10.9QNAKK458 pKa = 10.35KK459 pKa = 9.92EE460 pKa = 4.32LKK462 pKa = 10.35SRR464 pKa = 11.84LAKK467 pKa = 10.68ASVATVLAAQPVTTNLLKK485 pKa = 10.7FMRR488 pKa = 11.84DD489 pKa = 3.15LTRR492 pKa = 11.84TIGASNLQNFIASMTQKK509 pKa = 9.35LTLMDD514 pKa = 3.89TSDD517 pKa = 3.28EE518 pKa = 4.41TKK520 pKa = 10.85NYY522 pKa = 9.64LAAAGITTACLRR534 pKa = 11.84IKK536 pKa = 10.66SKK538 pKa = 8.22WTAGGKK544 pKa = 9.39
MM1 pKa = 7.44SVEE4 pKa = 4.3LIKK7 pKa = 10.7PLVEE11 pKa = 3.86AAGMSTLTQSPNLRR25 pKa = 11.84IWKK28 pKa = 8.93QVNTRR33 pKa = 11.84VKK35 pKa = 10.51LLEE38 pKa = 4.88EE39 pKa = 4.62ILAHH43 pKa = 5.1WTNGIRR49 pKa = 11.84RR50 pKa = 11.84CPEE53 pKa = 3.56TQDD56 pKa = 4.78FYY58 pKa = 11.45MNPNSQLATTIAYY71 pKa = 8.04RR72 pKa = 11.84AHH74 pKa = 6.19QKK76 pKa = 9.51GHH78 pKa = 5.39NPKK81 pKa = 10.28LIQYY85 pKa = 8.27PDD87 pKa = 3.65TFTLDD92 pKa = 3.71DD93 pKa = 4.6IIRR96 pKa = 11.84RR97 pKa = 11.84KK98 pKa = 9.8PIPQTAPNEE107 pKa = 3.94LRR109 pKa = 11.84LSDD112 pKa = 5.03EE113 pKa = 3.69IGEE116 pKa = 4.37DD117 pKa = 3.51YY118 pKa = 10.75RR119 pKa = 11.84LTVIAIADD127 pKa = 4.11EE128 pKa = 4.23LQEE131 pKa = 4.7CYY133 pKa = 10.51DD134 pKa = 3.64VLGQLDD140 pKa = 3.83INNTIDD146 pKa = 3.4HH147 pKa = 6.15TPKK150 pKa = 10.95GNAHH154 pKa = 6.5WSMLYY159 pKa = 10.22DD160 pKa = 4.05KK161 pKa = 10.32PVNTHH166 pKa = 5.13WSQLVSKK173 pKa = 10.58RR174 pKa = 11.84PLKK177 pKa = 10.43DD178 pKa = 2.77IRR180 pKa = 11.84ADD182 pKa = 3.47YY183 pKa = 10.94NYY185 pKa = 11.09AKK187 pKa = 10.74AKK189 pKa = 9.85GIKK192 pKa = 9.69DD193 pKa = 3.59EE194 pKa = 4.13CSKK197 pKa = 10.4MLEE200 pKa = 4.05EE201 pKa = 4.36STMKK205 pKa = 10.09SRR207 pKa = 11.84RR208 pKa = 11.84GFTVQRR214 pKa = 11.84LMNAMRR220 pKa = 11.84QAHH223 pKa = 5.52TDD225 pKa = 2.56GWYY228 pKa = 10.64VVFDD232 pKa = 4.23TLTLADD238 pKa = 4.35DD239 pKa = 5.61RR240 pKa = 11.84IQAFYY245 pKa = 11.18DD246 pKa = 3.46NPNALRR252 pKa = 11.84DD253 pKa = 3.67YY254 pKa = 10.67FRR256 pKa = 11.84DD257 pKa = 3.04IGRR260 pKa = 11.84MVLAAEE266 pKa = 4.55GRR268 pKa = 11.84SANDD272 pKa = 3.16SHH274 pKa = 7.55SDD276 pKa = 3.48CYY278 pKa = 11.08QYY280 pKa = 11.34FCVPEE285 pKa = 4.02YY286 pKa = 9.72GTQHH290 pKa = 5.68GRR292 pKa = 11.84LHH294 pKa = 5.96FHH296 pKa = 6.87AVHH299 pKa = 6.99LMRR302 pKa = 11.84TLPLGSRR309 pKa = 11.84DD310 pKa = 3.68PNFGKK315 pKa = 10.52LVRR318 pKa = 11.84TNRR321 pKa = 11.84QINSLQNTWPYY332 pKa = 9.75GYY334 pKa = 10.78SMPIAVRR341 pKa = 11.84YY342 pKa = 9.26SQDD345 pKa = 2.82AFSRR349 pKa = 11.84AGWLWPVDD357 pKa = 4.15SKK359 pKa = 11.92GEE361 pKa = 4.01PLKK364 pKa = 10.22ATSYY368 pKa = 9.01MAVGFYY374 pKa = 9.16VAKK377 pKa = 10.34YY378 pKa = 9.45VNKK381 pKa = 10.55KK382 pKa = 10.41SDD384 pKa = 2.79IDD386 pKa = 3.67MAAKK390 pKa = 10.6GLGNKK395 pKa = 7.49EE396 pKa = 3.68WNASLKK402 pKa = 9.91TKK404 pKa = 10.52ISLLPTKK411 pKa = 10.29LFRR414 pKa = 11.84IRR416 pKa = 11.84MSRR419 pKa = 11.84NFGMKK424 pKa = 8.53MLSMAHH430 pKa = 5.91LTAEE434 pKa = 4.48ALIQLTQLGYY444 pKa = 11.13DD445 pKa = 3.19STPFNNVLKK454 pKa = 10.9QNAKK458 pKa = 10.35KK459 pKa = 9.92EE460 pKa = 4.32LKK462 pKa = 10.35SRR464 pKa = 11.84LAKK467 pKa = 10.68ASVATVLAAQPVTTNLLKK485 pKa = 10.7FMRR488 pKa = 11.84DD489 pKa = 3.15LTRR492 pKa = 11.84TIGASNLQNFIASMTQKK509 pKa = 9.35LTLMDD514 pKa = 3.89TSDD517 pKa = 3.28EE518 pKa = 4.41TKK520 pKa = 10.85NYY522 pKa = 9.64LAAAGITTACLRR534 pKa = 11.84IKK536 pKa = 10.66SKK538 pKa = 8.22WTAGGKK544 pKa = 9.39
Molecular weight: 61.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1728 |
86 |
544 |
288.0 |
31.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.722 ± 0.932 | 0.868 ± 0.191 |
5.845 ± 0.322 | 3.704 ± 0.598 |
3.704 ± 0.499 | 6.481 ± 0.806 |
2.083 ± 0.559 | 5.382 ± 0.434 |
5.498 ± 0.925 | 8.275 ± 0.932 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.951 ± 0.42 | 5.382 ± 0.283 |
4.225 ± 0.478 | 5.093 ± 0.705 |
5.093 ± 0.707 | 7.292 ± 0.758 |
7.813 ± 0.843 | 5.729 ± 0.861 |
1.331 ± 0.27 | 3.53 ± 0.596 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
