
Pseudorhodoplanes sinuspersici
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiales incertae sedis; Pseudorhodoplanes
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
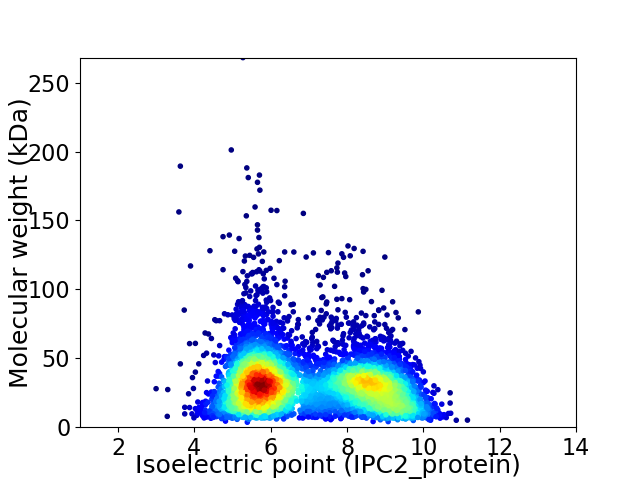
Virtual 2D-PAGE plot for 5631 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
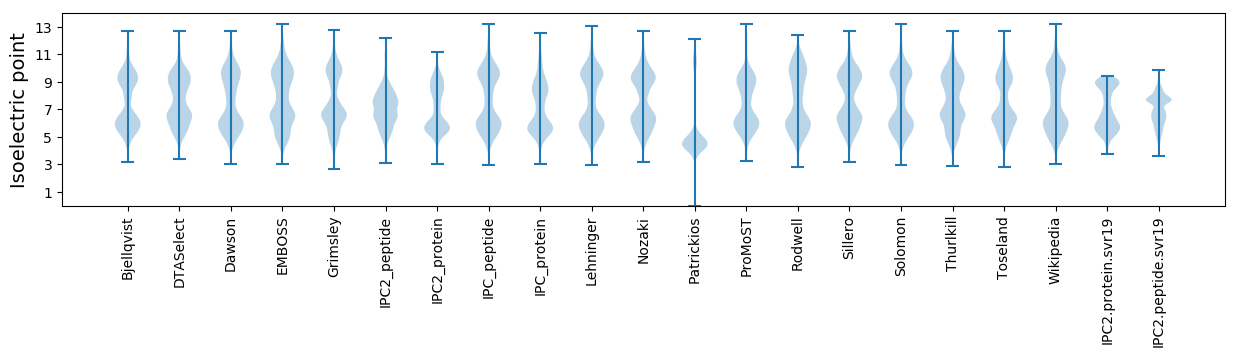
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6ZWH1|A0A1W6ZWH1_9RHIZ Protein-export membrane protein SecG OS=Pseudorhodoplanes sinuspersici OX=1235591 GN=CAK95_23085 PE=3 SV=1
MM1 pKa = 6.96QNARR5 pKa = 11.84FEE7 pKa = 4.83SVNTAATLRR16 pKa = 11.84TSSTIVQPSAATLEE30 pKa = 3.84NGILYY35 pKa = 7.6GTNRR39 pKa = 11.84DD40 pKa = 3.55DD41 pKa = 5.0VLFTTPQYY49 pKa = 10.71PHH51 pKa = 7.18LAGLTGADD59 pKa = 3.59RR60 pKa = 11.84LTGDD64 pKa = 3.27EE65 pKa = 4.49RR66 pKa = 11.84DD67 pKa = 3.26NRR69 pKa = 11.84LYY71 pKa = 11.09GGAHH75 pKa = 6.45NDD77 pKa = 3.34QLFGQDD83 pKa = 4.18GNDD86 pKa = 3.24ILDD89 pKa = 4.17GGTEE93 pKa = 3.99ADD95 pKa = 4.12RR96 pKa = 11.84LEE98 pKa = 4.65GGAGNDD104 pKa = 3.02IYY106 pKa = 11.52YY107 pKa = 10.25VDD109 pKa = 4.5HH110 pKa = 6.57FQDD113 pKa = 3.91VIIEE117 pKa = 4.2EE118 pKa = 4.28ADD120 pKa = 3.11GGYY123 pKa = 10.65DD124 pKa = 4.4IMFTNPDD131 pKa = 3.67GIYY134 pKa = 10.59GNSHH138 pKa = 6.0RR139 pKa = 11.84MALNVEE145 pKa = 4.1QMIMTGSGWQMGEE158 pKa = 4.8GNTQDD163 pKa = 3.16NFIIGSEE170 pKa = 4.17SEE172 pKa = 4.18NLILAYY178 pKa = 10.33GGDD181 pKa = 4.12DD182 pKa = 2.58IVYY185 pKa = 10.48GMGGTDD191 pKa = 4.41RR192 pKa = 11.84IYY194 pKa = 11.21GDD196 pKa = 5.0DD197 pKa = 4.13LNAPYY202 pKa = 10.51AGNDD206 pKa = 2.91ILYY209 pKa = 10.29GGEE212 pKa = 4.34GDD214 pKa = 3.97DD215 pKa = 4.47FLYY218 pKa = 11.0GGLGDD223 pKa = 4.23DD224 pKa = 3.64MLYY227 pKa = 10.82GGRR230 pKa = 11.84GVDD233 pKa = 3.49MLSGSGGNDD242 pKa = 2.55TGVFADD248 pKa = 4.41AEE250 pKa = 4.46SAVIIDD256 pKa = 3.71LTRR259 pKa = 11.84PYY261 pKa = 11.13NNGGSATGEE270 pKa = 3.85YY271 pKa = 10.45LFSIEE276 pKa = 4.36NLIGSAFNDD285 pKa = 4.04TLSGGQSRR293 pKa = 11.84NMLEE297 pKa = 4.43GGDD300 pKa = 3.83GNDD303 pKa = 2.93RR304 pKa = 11.84LFGRR308 pKa = 11.84EE309 pKa = 3.6GGDD312 pKa = 3.45TVRR315 pKa = 11.84GGAGSDD321 pKa = 4.13FIDD324 pKa = 4.03GGANRR329 pKa = 11.84DD330 pKa = 3.75VLTGGAGIGDD340 pKa = 4.27APDD343 pKa = 3.26TFYY346 pKa = 10.92FASAAEE352 pKa = 4.42AGDD355 pKa = 3.94TITDD359 pKa = 4.58FFSDD363 pKa = 4.66HH364 pKa = 6.59IALSAEE370 pKa = 4.07GFGLEE375 pKa = 4.41SVDD378 pKa = 3.92DD379 pKa = 4.04FVFARR384 pKa = 11.84TSDD387 pKa = 3.97PLTDD391 pKa = 4.32MPTMIYY397 pKa = 10.5DD398 pKa = 3.63SRR400 pKa = 11.84SGNLSWDD407 pKa = 3.42ADD409 pKa = 3.77GSGGEE414 pKa = 3.94AAVYY418 pKa = 9.09FATLTGAPLLTQNDD432 pKa = 3.77FLIII436 pKa = 3.93
MM1 pKa = 6.96QNARR5 pKa = 11.84FEE7 pKa = 4.83SVNTAATLRR16 pKa = 11.84TSSTIVQPSAATLEE30 pKa = 3.84NGILYY35 pKa = 7.6GTNRR39 pKa = 11.84DD40 pKa = 3.55DD41 pKa = 5.0VLFTTPQYY49 pKa = 10.71PHH51 pKa = 7.18LAGLTGADD59 pKa = 3.59RR60 pKa = 11.84LTGDD64 pKa = 3.27EE65 pKa = 4.49RR66 pKa = 11.84DD67 pKa = 3.26NRR69 pKa = 11.84LYY71 pKa = 11.09GGAHH75 pKa = 6.45NDD77 pKa = 3.34QLFGQDD83 pKa = 4.18GNDD86 pKa = 3.24ILDD89 pKa = 4.17GGTEE93 pKa = 3.99ADD95 pKa = 4.12RR96 pKa = 11.84LEE98 pKa = 4.65GGAGNDD104 pKa = 3.02IYY106 pKa = 11.52YY107 pKa = 10.25VDD109 pKa = 4.5HH110 pKa = 6.57FQDD113 pKa = 3.91VIIEE117 pKa = 4.2EE118 pKa = 4.28ADD120 pKa = 3.11GGYY123 pKa = 10.65DD124 pKa = 4.4IMFTNPDD131 pKa = 3.67GIYY134 pKa = 10.59GNSHH138 pKa = 6.0RR139 pKa = 11.84MALNVEE145 pKa = 4.1QMIMTGSGWQMGEE158 pKa = 4.8GNTQDD163 pKa = 3.16NFIIGSEE170 pKa = 4.17SEE172 pKa = 4.18NLILAYY178 pKa = 10.33GGDD181 pKa = 4.12DD182 pKa = 2.58IVYY185 pKa = 10.48GMGGTDD191 pKa = 4.41RR192 pKa = 11.84IYY194 pKa = 11.21GDD196 pKa = 5.0DD197 pKa = 4.13LNAPYY202 pKa = 10.51AGNDD206 pKa = 2.91ILYY209 pKa = 10.29GGEE212 pKa = 4.34GDD214 pKa = 3.97DD215 pKa = 4.47FLYY218 pKa = 11.0GGLGDD223 pKa = 4.23DD224 pKa = 3.64MLYY227 pKa = 10.82GGRR230 pKa = 11.84GVDD233 pKa = 3.49MLSGSGGNDD242 pKa = 2.55TGVFADD248 pKa = 4.41AEE250 pKa = 4.46SAVIIDD256 pKa = 3.71LTRR259 pKa = 11.84PYY261 pKa = 11.13NNGGSATGEE270 pKa = 3.85YY271 pKa = 10.45LFSIEE276 pKa = 4.36NLIGSAFNDD285 pKa = 4.04TLSGGQSRR293 pKa = 11.84NMLEE297 pKa = 4.43GGDD300 pKa = 3.83GNDD303 pKa = 2.93RR304 pKa = 11.84LFGRR308 pKa = 11.84EE309 pKa = 3.6GGDD312 pKa = 3.45TVRR315 pKa = 11.84GGAGSDD321 pKa = 4.13FIDD324 pKa = 4.03GGANRR329 pKa = 11.84DD330 pKa = 3.75VLTGGAGIGDD340 pKa = 4.27APDD343 pKa = 3.26TFYY346 pKa = 10.92FASAAEE352 pKa = 4.42AGDD355 pKa = 3.94TITDD359 pKa = 4.58FFSDD363 pKa = 4.66HH364 pKa = 6.59IALSAEE370 pKa = 4.07GFGLEE375 pKa = 4.41SVDD378 pKa = 3.92DD379 pKa = 4.04FVFARR384 pKa = 11.84TSDD387 pKa = 3.97PLTDD391 pKa = 4.32MPTMIYY397 pKa = 10.5DD398 pKa = 3.63SRR400 pKa = 11.84SGNLSWDD407 pKa = 3.42ADD409 pKa = 3.77GSGGEE414 pKa = 3.94AAVYY418 pKa = 9.09FATLTGAPLLTQNDD432 pKa = 3.77FLIII436 pKa = 3.93
Molecular weight: 45.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6ZRP6|A0A1W6ZRP6_9RHIZ Gamma-glutamyltransferase OS=Pseudorhodoplanes sinuspersici OX=1235591 GN=CAK95_10305 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1729092 |
31 |
2510 |
307.1 |
33.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.984 ± 0.039 | 0.842 ± 0.011 |
5.627 ± 0.028 | 5.282 ± 0.029 |
3.874 ± 0.023 | 8.214 ± 0.029 |
2.044 ± 0.016 | 5.678 ± 0.023 |
3.894 ± 0.027 | 9.746 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.565 ± 0.016 | 2.843 ± 0.02 |
5.305 ± 0.025 | 3.181 ± 0.017 |
6.922 ± 0.031 | 5.496 ± 0.021 |
5.395 ± 0.024 | 7.453 ± 0.023 |
1.321 ± 0.014 | 2.335 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
