
Sorangium cellulosum (strain So ce56) (Polyangium cellulosum (strain So ce56))
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Sorangiineae; Polyangiaceae; Sorangium; Sorangium cellulosum
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
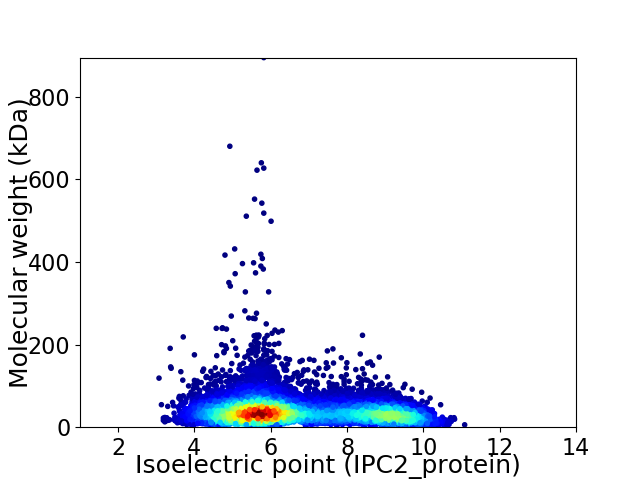
Virtual 2D-PAGE plot for 9320 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
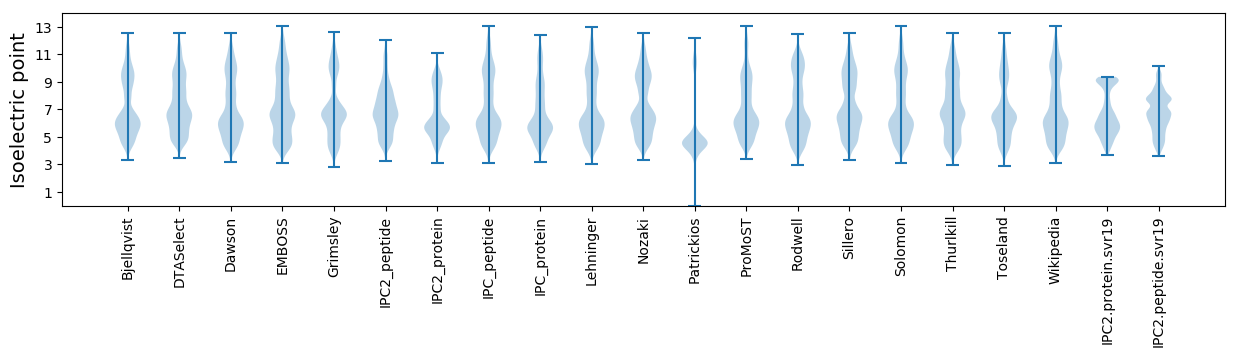
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A9GAX1|A9GAX1_SORC5 Uncharacterized protein OS=Sorangium cellulosum (strain So ce56) OX=448385 GN=sce2764 PE=4 SV=1
MM1 pKa = 7.56RR2 pKa = 11.84KK3 pKa = 8.96SWLAVMFSVASMACLTSGCSALPDD27 pKa = 3.89GDD29 pKa = 4.48DD30 pKa = 3.8PRR32 pKa = 11.84TSEE35 pKa = 4.58PVGTSQQPLVYY46 pKa = 9.54GWSNRR51 pKa = 11.84YY52 pKa = 9.46GDD54 pKa = 4.27INYY57 pKa = 8.92QHH59 pKa = 7.03AGDD62 pKa = 3.93VTVDD66 pKa = 3.22VSDD69 pKa = 5.05NIIITAGGEE78 pKa = 4.14GSVDD82 pKa = 4.03FGGGAHH88 pKa = 6.28TPTFYY93 pKa = 11.47GVMLAKK99 pKa = 10.27FDD101 pKa = 5.42DD102 pKa = 4.28SGNCIWSKK110 pKa = 9.81MFEE113 pKa = 4.48GVNLMGVATDD123 pKa = 3.36SAGNVNIVGTMFGAIDD139 pKa = 3.87FGGGPMSPLGMSDD152 pKa = 2.85IFVAQFDD159 pKa = 3.71ASGNYY164 pKa = 7.59VWSDD168 pKa = 3.17RR169 pKa = 11.84FGGGPLEE176 pKa = 4.37YY177 pKa = 10.8AEE179 pKa = 4.52GMAIAVDD186 pKa = 3.62SSDD189 pKa = 3.44NVIITGNYY197 pKa = 8.98SGNLRR202 pKa = 11.84FGSDD206 pKa = 3.07FGIAVSNDD214 pKa = 2.74VFLAKK219 pKa = 10.03LDD221 pKa = 3.83SSGNPVFGRR230 pKa = 11.84FFGVPLVPGTGTGRR244 pKa = 11.84AVAIDD249 pKa = 3.89GSDD252 pKa = 3.69NIVISGWYY260 pKa = 8.21YY261 pKa = 10.85GSMDD265 pKa = 3.99FGGGPTPLVSVPDD278 pKa = 3.67TADD281 pKa = 3.12AFVAEE286 pKa = 4.85FDD288 pKa = 4.49SSGTYY293 pKa = 10.3LWDD296 pKa = 3.42TTFGNSGEE304 pKa = 4.04ADD306 pKa = 3.73ALNVATDD313 pKa = 3.41SLGNVAVVGSFTGNMDD329 pKa = 2.89IGGSTLTSAGNTDD342 pKa = 2.85IFVAQFDD349 pKa = 3.85NSGTHH354 pKa = 5.84LWSHH358 pKa = 6.27GFGDD362 pKa = 4.55TDD364 pKa = 3.11WDD366 pKa = 3.8MASGVSINDD375 pKa = 3.39SGDD378 pKa = 3.31VFVTGDD384 pKa = 2.75IGGAVDD390 pKa = 4.7FGGGTLTSAGSSDD403 pKa = 3.6VFVAHH408 pKa = 6.81FDD410 pKa = 3.59GSGTHH415 pKa = 5.77VWSEE419 pKa = 4.23NYY421 pKa = 10.62GDD423 pKa = 4.26TDD425 pKa = 3.77PQGGSSIKK433 pKa = 9.82IDD435 pKa = 3.66SNGNAVVTGAFFGSVNFGGTTLTDD459 pKa = 3.47AGTGDD464 pKa = 3.03IFLAQFTPP472 pKa = 3.74
MM1 pKa = 7.56RR2 pKa = 11.84KK3 pKa = 8.96SWLAVMFSVASMACLTSGCSALPDD27 pKa = 3.89GDD29 pKa = 4.48DD30 pKa = 3.8PRR32 pKa = 11.84TSEE35 pKa = 4.58PVGTSQQPLVYY46 pKa = 9.54GWSNRR51 pKa = 11.84YY52 pKa = 9.46GDD54 pKa = 4.27INYY57 pKa = 8.92QHH59 pKa = 7.03AGDD62 pKa = 3.93VTVDD66 pKa = 3.22VSDD69 pKa = 5.05NIIITAGGEE78 pKa = 4.14GSVDD82 pKa = 4.03FGGGAHH88 pKa = 6.28TPTFYY93 pKa = 11.47GVMLAKK99 pKa = 10.27FDD101 pKa = 5.42DD102 pKa = 4.28SGNCIWSKK110 pKa = 9.81MFEE113 pKa = 4.48GVNLMGVATDD123 pKa = 3.36SAGNVNIVGTMFGAIDD139 pKa = 3.87FGGGPMSPLGMSDD152 pKa = 2.85IFVAQFDD159 pKa = 3.71ASGNYY164 pKa = 7.59VWSDD168 pKa = 3.17RR169 pKa = 11.84FGGGPLEE176 pKa = 4.37YY177 pKa = 10.8AEE179 pKa = 4.52GMAIAVDD186 pKa = 3.62SSDD189 pKa = 3.44NVIITGNYY197 pKa = 8.98SGNLRR202 pKa = 11.84FGSDD206 pKa = 3.07FGIAVSNDD214 pKa = 2.74VFLAKK219 pKa = 10.03LDD221 pKa = 3.83SSGNPVFGRR230 pKa = 11.84FFGVPLVPGTGTGRR244 pKa = 11.84AVAIDD249 pKa = 3.89GSDD252 pKa = 3.69NIVISGWYY260 pKa = 8.21YY261 pKa = 10.85GSMDD265 pKa = 3.99FGGGPTPLVSVPDD278 pKa = 3.67TADD281 pKa = 3.12AFVAEE286 pKa = 4.85FDD288 pKa = 4.49SSGTYY293 pKa = 10.3LWDD296 pKa = 3.42TTFGNSGEE304 pKa = 4.04ADD306 pKa = 3.73ALNVATDD313 pKa = 3.41SLGNVAVVGSFTGNMDD329 pKa = 2.89IGGSTLTSAGNTDD342 pKa = 2.85IFVAQFDD349 pKa = 3.85NSGTHH354 pKa = 5.84LWSHH358 pKa = 6.27GFGDD362 pKa = 4.55TDD364 pKa = 3.11WDD366 pKa = 3.8MASGVSINDD375 pKa = 3.39SGDD378 pKa = 3.31VFVTGDD384 pKa = 2.75IGGAVDD390 pKa = 4.7FGGGTLTSAGSSDD403 pKa = 3.6VFVAHH408 pKa = 6.81FDD410 pKa = 3.59GSGTHH415 pKa = 5.77VWSEE419 pKa = 4.23NYY421 pKa = 10.62GDD423 pKa = 4.26TDD425 pKa = 3.77PQGGSSIKK433 pKa = 9.82IDD435 pKa = 3.66SNGNAVVTGAFFGSVNFGGTTLTDD459 pKa = 3.47AGTGDD464 pKa = 3.03IFLAQFTPP472 pKa = 3.74
Molecular weight: 48.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A9FTL4|A9FTL4_SORC5 CENP-V/GFA domain-containing protein OS=Sorangium cellulosum (strain So ce56) OX=448385 GN=sce8373 PE=3 SV=1
MM1 pKa = 6.99SHH3 pKa = 6.76ARR5 pKa = 11.84PVWSRR10 pKa = 11.84HH11 pKa = 4.45RR12 pKa = 11.84SAPGPGTRR20 pKa = 11.84SRR22 pKa = 11.84IHH24 pKa = 6.01TRR26 pKa = 11.84DD27 pKa = 3.29ASEE30 pKa = 4.01QGPPATSTPVGWQGRR45 pKa = 11.84LAGPPQPRR53 pKa = 11.84SGWLAGPGPPQSRR66 pKa = 11.84SGPPNLGRR74 pKa = 11.84VRR76 pKa = 11.84LNLGRR81 pKa = 11.84VRR83 pKa = 11.84LNLGRR88 pKa = 11.84VRR90 pKa = 11.84LNLGRR95 pKa = 11.84VRR97 pKa = 11.84LNLGRR102 pKa = 11.84VRR104 pKa = 11.84LSLGRR109 pKa = 11.84GRR111 pKa = 11.84LNLGRR116 pKa = 11.84GRR118 pKa = 11.84LNLGRR123 pKa = 11.84VRR125 pKa = 11.84LNLGRR130 pKa = 11.84VRR132 pKa = 11.84LNLGRR137 pKa = 11.84VRR139 pKa = 11.84LNLGRR144 pKa = 11.84VHH146 pKa = 7.5PARR149 pKa = 11.84ASAVAGGAAALTCAAGAPIPPHH171 pKa = 5.61LQRR174 pKa = 11.84TAARR178 pKa = 11.84TGGSRR183 pKa = 11.84RR184 pKa = 11.84PPSPLDD190 pKa = 3.53LSPEE194 pKa = 4.01LSDD197 pKa = 3.77ALARR201 pKa = 11.84SIPLPPTEE209 pKa = 4.19PWRR212 pKa = 11.84KK213 pKa = 9.4RR214 pKa = 11.84SS215 pKa = 3.38
MM1 pKa = 6.99SHH3 pKa = 6.76ARR5 pKa = 11.84PVWSRR10 pKa = 11.84HH11 pKa = 4.45RR12 pKa = 11.84SAPGPGTRR20 pKa = 11.84SRR22 pKa = 11.84IHH24 pKa = 6.01TRR26 pKa = 11.84DD27 pKa = 3.29ASEE30 pKa = 4.01QGPPATSTPVGWQGRR45 pKa = 11.84LAGPPQPRR53 pKa = 11.84SGWLAGPGPPQSRR66 pKa = 11.84SGPPNLGRR74 pKa = 11.84VRR76 pKa = 11.84LNLGRR81 pKa = 11.84VRR83 pKa = 11.84LNLGRR88 pKa = 11.84VRR90 pKa = 11.84LNLGRR95 pKa = 11.84VRR97 pKa = 11.84LNLGRR102 pKa = 11.84VRR104 pKa = 11.84LSLGRR109 pKa = 11.84GRR111 pKa = 11.84LNLGRR116 pKa = 11.84GRR118 pKa = 11.84LNLGRR123 pKa = 11.84VRR125 pKa = 11.84LNLGRR130 pKa = 11.84VRR132 pKa = 11.84LNLGRR137 pKa = 11.84VRR139 pKa = 11.84LNLGRR144 pKa = 11.84VHH146 pKa = 7.5PARR149 pKa = 11.84ASAVAGGAAALTCAAGAPIPPHH171 pKa = 5.61LQRR174 pKa = 11.84TAARR178 pKa = 11.84TGGSRR183 pKa = 11.84RR184 pKa = 11.84PPSPLDD190 pKa = 3.53LSPEE194 pKa = 4.01LSDD197 pKa = 3.77ALARR201 pKa = 11.84SIPLPPTEE209 pKa = 4.19PWRR212 pKa = 11.84KK213 pKa = 9.4RR214 pKa = 11.84SS215 pKa = 3.38
Molecular weight: 23.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3743060 |
30 |
8417 |
401.6 |
42.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.161 ± 0.046 | 1.185 ± 0.019 |
5.674 ± 0.022 | 6.237 ± 0.022 |
3.041 ± 0.014 | 9.362 ± 0.034 |
2.044 ± 0.011 | 3.601 ± 0.015 |
2.334 ± 0.019 | 10.131 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.702 ± 0.01 | 1.759 ± 0.015 |
6.369 ± 0.025 | 2.677 ± 0.018 |
8.625 ± 0.03 | 5.707 ± 0.019 |
4.753 ± 0.021 | 7.449 ± 0.022 |
1.269 ± 0.011 | 1.92 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
