
Wenling narna-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.43
Get precalculated fractions of proteins
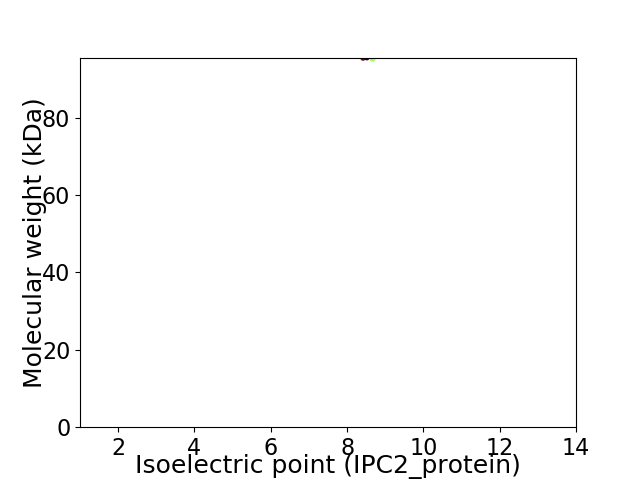
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
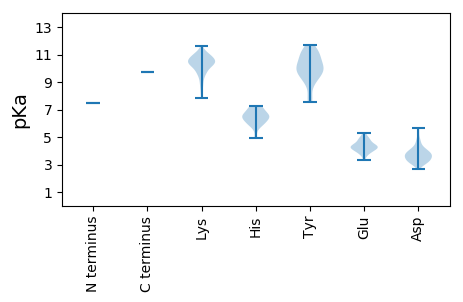
Protein with the lowest isoelectric point:
>tr|A0A1L3KIS0|A0A1L3KIS0_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 9 OX=1923509 PE=4 SV=1
MM1 pKa = 7.45ICILLMIMHH10 pKa = 7.26ILICVSRR17 pKa = 11.84SVQKK21 pKa = 10.85HH22 pKa = 4.95FFWSKK27 pKa = 9.73ANSWSAALKK36 pKa = 10.85NGDD39 pKa = 3.12GKK41 pKa = 11.12APNFDD46 pKa = 3.13RR47 pKa = 11.84KK48 pKa = 9.62MNQIVLSKK56 pKa = 9.58TYY58 pKa = 10.1KK59 pKa = 10.26ILMKK63 pKa = 10.45LSMGGVVNGILKK75 pKa = 10.36DD76 pKa = 3.1IYY78 pKa = 10.18TSHH81 pKa = 6.14NKK83 pKa = 9.92KK84 pKa = 10.28LEE86 pKa = 4.26KK87 pKa = 10.6YY88 pKa = 9.28SANAVIQQFKK98 pKa = 10.93EE99 pKa = 3.73DD100 pKa = 3.98FLFVKK105 pKa = 10.52NYY107 pKa = 10.26FLMGTVSEE115 pKa = 5.01FDD117 pKa = 4.17QKK119 pKa = 11.1LQWRR123 pKa = 11.84RR124 pKa = 11.84LSDD127 pKa = 3.19VGTFSLPPYY136 pKa = 9.88LSPIYY141 pKa = 10.26HH142 pKa = 7.27RR143 pKa = 11.84LGEE146 pKa = 4.21LNSWDD151 pKa = 3.72KK152 pKa = 11.59VLLLTLLRR160 pKa = 11.84SYY162 pKa = 11.29KK163 pKa = 10.75HH164 pKa = 5.75MDD166 pKa = 2.69IPGPHH171 pKa = 7.09KK172 pKa = 10.95LEE174 pKa = 4.64TIYY177 pKa = 11.21TMMGYY182 pKa = 7.68QTYY185 pKa = 9.64INTYY189 pKa = 7.83KK190 pKa = 10.4VCMSMSCVYY199 pKa = 10.71KK200 pKa = 10.34WMGMPKK206 pKa = 9.99LQPSDD211 pKa = 3.17TGDD214 pKa = 3.35YY215 pKa = 11.01SKK217 pKa = 11.04ISIAKK222 pKa = 9.68GLTPGTNTGSLLHH235 pKa = 6.61AVGPSGNTLVSRR247 pKa = 11.84LVDD250 pKa = 4.02LMNVYY255 pKa = 9.86QKK257 pKa = 11.12PMYY260 pKa = 10.56AGLLDD265 pKa = 4.73LFAKK269 pKa = 10.31FYY271 pKa = 11.3SDD273 pKa = 3.34FTKK276 pKa = 10.57VGKK279 pKa = 10.7GPGITNVSKK288 pKa = 10.95LFQHH292 pKa = 6.55FSKK295 pKa = 10.7PVLDD299 pKa = 4.29EE300 pKa = 4.09TLSMAKK306 pKa = 9.62EE307 pKa = 3.83NSKK310 pKa = 10.4IKK312 pKa = 10.36PKK314 pKa = 10.67VDD316 pKa = 3.18MKK318 pKa = 10.72ILNRR322 pKa = 11.84LSYY325 pKa = 9.47FTDD328 pKa = 4.29GAGKK332 pKa = 8.65WRR334 pKa = 11.84YY335 pKa = 8.9IAISDD340 pKa = 4.74WITQSILSPMHH351 pKa = 6.46NLMFDD356 pKa = 3.58ALKK359 pKa = 11.1SMSTDD364 pKa = 3.23CTFDD368 pKa = 3.3QNKK371 pKa = 8.84IRR373 pKa = 11.84AKK375 pKa = 7.85YY376 pKa = 9.79QEE378 pKa = 4.88MYY380 pKa = 10.46SQGKK384 pKa = 8.53GAYY387 pKa = 9.8CLDD390 pKa = 3.97LSAATDD396 pKa = 3.9RR397 pKa = 11.84LPLPLQAFLMFLLTKK412 pKa = 10.73NFGLSLSWMLIMMYY426 pKa = 10.6NPFHH430 pKa = 6.46LTGDD434 pKa = 3.81MEE436 pKa = 4.74KK437 pKa = 10.62KK438 pKa = 10.54FYY440 pKa = 10.34YY441 pKa = 9.05QTGQGMGIYY450 pKa = 9.4TSWAMLALTNHH461 pKa = 7.03AIVRR465 pKa = 11.84LSGKK469 pKa = 10.33VYY471 pKa = 9.94GHH473 pKa = 6.61SDD475 pKa = 3.18FSDD478 pKa = 3.76YY479 pKa = 11.3LVLGDD484 pKa = 5.29DD485 pKa = 3.53VVIFNKK491 pKa = 9.92EE492 pKa = 3.8VADD495 pKa = 4.7GYY497 pKa = 9.32LTVMSKK503 pKa = 10.87LGVDD507 pKa = 3.65ISQPKK512 pKa = 10.43SIFPKK517 pKa = 9.9EE518 pKa = 3.98NKK520 pKa = 9.57FGCEE524 pKa = 3.32FASRR528 pKa = 11.84LFVEE532 pKa = 4.89GQEE535 pKa = 4.31LSPLPVGLLIKK546 pKa = 10.44DD547 pKa = 3.8PNVDD551 pKa = 3.61NILNFWTTVHH561 pKa = 6.1WKK563 pKa = 8.19STEE566 pKa = 4.01LVEE569 pKa = 4.42TQYY572 pKa = 11.35QDD574 pKa = 4.09SLTTPDD580 pKa = 4.2LSSAVPLSGSEE591 pKa = 4.02DD592 pKa = 3.63LKK594 pKa = 10.35TIWGFMSVYY603 pKa = 10.09NYY605 pKa = 10.95YY606 pKa = 8.72YY607 pKa = 7.58TQHH610 pKa = 5.96MKK612 pKa = 10.43FSEE615 pKa = 4.21NEE617 pKa = 4.19VPSGAWMSLPEE628 pKa = 4.56ASPLHH633 pKa = 5.88TLLEE637 pKa = 4.81AIPLKK642 pKa = 10.98SMEE645 pKa = 4.32EE646 pKa = 4.16VEE648 pKa = 4.33STMEE652 pKa = 4.3KK653 pKa = 9.22ILYY656 pKa = 9.9RR657 pKa = 11.84RR658 pKa = 11.84FSQALKK664 pKa = 10.35TMTKK668 pKa = 9.89HH669 pKa = 6.58SKK671 pKa = 9.18TFDD674 pKa = 3.62SIKK677 pKa = 11.22DD678 pKa = 3.58MLVPLYY684 pKa = 10.85GSLGDD689 pKa = 3.74LEE691 pKa = 5.3SRR693 pKa = 11.84FPHH696 pKa = 6.46ILSMYY701 pKa = 9.55LFFMSPFYY709 pKa = 11.05HH710 pKa = 6.99CIDD713 pKa = 3.87KK714 pKa = 10.79FSEE717 pKa = 4.33LNLDD721 pKa = 3.9PQSEE725 pKa = 4.48MDD727 pKa = 3.31SHH729 pKa = 6.65MIAAGSGVLVMDD741 pKa = 4.01QAQKK745 pKa = 10.33LDD747 pKa = 3.35HH748 pKa = 6.09MAKK751 pKa = 10.28FFTFAPLSMHH761 pKa = 6.38KK762 pKa = 10.49KK763 pKa = 9.64IDD765 pKa = 3.54IEE767 pKa = 4.35SLSDD771 pKa = 3.69LLKK774 pKa = 10.46ISYY777 pKa = 9.74GPKK780 pKa = 10.1SLFLGKK786 pKa = 10.06DD787 pKa = 3.04DD788 pKa = 5.68WILKK792 pKa = 10.51DD793 pKa = 3.67PLTKK797 pKa = 10.2KK798 pKa = 10.43VLSKK802 pKa = 10.77KK803 pKa = 9.05VLSRR807 pKa = 11.84QRR809 pKa = 11.84VSKK812 pKa = 10.84KK813 pKa = 10.17DD814 pKa = 3.27VQNAIRR820 pKa = 11.84VSTFRR825 pKa = 11.84MAVMDD830 pKa = 3.59QYY832 pKa = 11.71LRR834 pKa = 11.84SNKK837 pKa = 9.76
MM1 pKa = 7.45ICILLMIMHH10 pKa = 7.26ILICVSRR17 pKa = 11.84SVQKK21 pKa = 10.85HH22 pKa = 4.95FFWSKK27 pKa = 9.73ANSWSAALKK36 pKa = 10.85NGDD39 pKa = 3.12GKK41 pKa = 11.12APNFDD46 pKa = 3.13RR47 pKa = 11.84KK48 pKa = 9.62MNQIVLSKK56 pKa = 9.58TYY58 pKa = 10.1KK59 pKa = 10.26ILMKK63 pKa = 10.45LSMGGVVNGILKK75 pKa = 10.36DD76 pKa = 3.1IYY78 pKa = 10.18TSHH81 pKa = 6.14NKK83 pKa = 9.92KK84 pKa = 10.28LEE86 pKa = 4.26KK87 pKa = 10.6YY88 pKa = 9.28SANAVIQQFKK98 pKa = 10.93EE99 pKa = 3.73DD100 pKa = 3.98FLFVKK105 pKa = 10.52NYY107 pKa = 10.26FLMGTVSEE115 pKa = 5.01FDD117 pKa = 4.17QKK119 pKa = 11.1LQWRR123 pKa = 11.84RR124 pKa = 11.84LSDD127 pKa = 3.19VGTFSLPPYY136 pKa = 9.88LSPIYY141 pKa = 10.26HH142 pKa = 7.27RR143 pKa = 11.84LGEE146 pKa = 4.21LNSWDD151 pKa = 3.72KK152 pKa = 11.59VLLLTLLRR160 pKa = 11.84SYY162 pKa = 11.29KK163 pKa = 10.75HH164 pKa = 5.75MDD166 pKa = 2.69IPGPHH171 pKa = 7.09KK172 pKa = 10.95LEE174 pKa = 4.64TIYY177 pKa = 11.21TMMGYY182 pKa = 7.68QTYY185 pKa = 9.64INTYY189 pKa = 7.83KK190 pKa = 10.4VCMSMSCVYY199 pKa = 10.71KK200 pKa = 10.34WMGMPKK206 pKa = 9.99LQPSDD211 pKa = 3.17TGDD214 pKa = 3.35YY215 pKa = 11.01SKK217 pKa = 11.04ISIAKK222 pKa = 9.68GLTPGTNTGSLLHH235 pKa = 6.61AVGPSGNTLVSRR247 pKa = 11.84LVDD250 pKa = 4.02LMNVYY255 pKa = 9.86QKK257 pKa = 11.12PMYY260 pKa = 10.56AGLLDD265 pKa = 4.73LFAKK269 pKa = 10.31FYY271 pKa = 11.3SDD273 pKa = 3.34FTKK276 pKa = 10.57VGKK279 pKa = 10.7GPGITNVSKK288 pKa = 10.95LFQHH292 pKa = 6.55FSKK295 pKa = 10.7PVLDD299 pKa = 4.29EE300 pKa = 4.09TLSMAKK306 pKa = 9.62EE307 pKa = 3.83NSKK310 pKa = 10.4IKK312 pKa = 10.36PKK314 pKa = 10.67VDD316 pKa = 3.18MKK318 pKa = 10.72ILNRR322 pKa = 11.84LSYY325 pKa = 9.47FTDD328 pKa = 4.29GAGKK332 pKa = 8.65WRR334 pKa = 11.84YY335 pKa = 8.9IAISDD340 pKa = 4.74WITQSILSPMHH351 pKa = 6.46NLMFDD356 pKa = 3.58ALKK359 pKa = 11.1SMSTDD364 pKa = 3.23CTFDD368 pKa = 3.3QNKK371 pKa = 8.84IRR373 pKa = 11.84AKK375 pKa = 7.85YY376 pKa = 9.79QEE378 pKa = 4.88MYY380 pKa = 10.46SQGKK384 pKa = 8.53GAYY387 pKa = 9.8CLDD390 pKa = 3.97LSAATDD396 pKa = 3.9RR397 pKa = 11.84LPLPLQAFLMFLLTKK412 pKa = 10.73NFGLSLSWMLIMMYY426 pKa = 10.6NPFHH430 pKa = 6.46LTGDD434 pKa = 3.81MEE436 pKa = 4.74KK437 pKa = 10.62KK438 pKa = 10.54FYY440 pKa = 10.34YY441 pKa = 9.05QTGQGMGIYY450 pKa = 9.4TSWAMLALTNHH461 pKa = 7.03AIVRR465 pKa = 11.84LSGKK469 pKa = 10.33VYY471 pKa = 9.94GHH473 pKa = 6.61SDD475 pKa = 3.18FSDD478 pKa = 3.76YY479 pKa = 11.3LVLGDD484 pKa = 5.29DD485 pKa = 3.53VVIFNKK491 pKa = 9.92EE492 pKa = 3.8VADD495 pKa = 4.7GYY497 pKa = 9.32LTVMSKK503 pKa = 10.87LGVDD507 pKa = 3.65ISQPKK512 pKa = 10.43SIFPKK517 pKa = 9.9EE518 pKa = 3.98NKK520 pKa = 9.57FGCEE524 pKa = 3.32FASRR528 pKa = 11.84LFVEE532 pKa = 4.89GQEE535 pKa = 4.31LSPLPVGLLIKK546 pKa = 10.44DD547 pKa = 3.8PNVDD551 pKa = 3.61NILNFWTTVHH561 pKa = 6.1WKK563 pKa = 8.19STEE566 pKa = 4.01LVEE569 pKa = 4.42TQYY572 pKa = 11.35QDD574 pKa = 4.09SLTTPDD580 pKa = 4.2LSSAVPLSGSEE591 pKa = 4.02DD592 pKa = 3.63LKK594 pKa = 10.35TIWGFMSVYY603 pKa = 10.09NYY605 pKa = 10.95YY606 pKa = 8.72YY607 pKa = 7.58TQHH610 pKa = 5.96MKK612 pKa = 10.43FSEE615 pKa = 4.21NEE617 pKa = 4.19VPSGAWMSLPEE628 pKa = 4.56ASPLHH633 pKa = 5.88TLLEE637 pKa = 4.81AIPLKK642 pKa = 10.98SMEE645 pKa = 4.32EE646 pKa = 4.16VEE648 pKa = 4.33STMEE652 pKa = 4.3KK653 pKa = 9.22ILYY656 pKa = 9.9RR657 pKa = 11.84RR658 pKa = 11.84FSQALKK664 pKa = 10.35TMTKK668 pKa = 9.89HH669 pKa = 6.58SKK671 pKa = 9.18TFDD674 pKa = 3.62SIKK677 pKa = 11.22DD678 pKa = 3.58MLVPLYY684 pKa = 10.85GSLGDD689 pKa = 3.74LEE691 pKa = 5.3SRR693 pKa = 11.84FPHH696 pKa = 6.46ILSMYY701 pKa = 9.55LFFMSPFYY709 pKa = 11.05HH710 pKa = 6.99CIDD713 pKa = 3.87KK714 pKa = 10.79FSEE717 pKa = 4.33LNLDD721 pKa = 3.9PQSEE725 pKa = 4.48MDD727 pKa = 3.31SHH729 pKa = 6.65MIAAGSGVLVMDD741 pKa = 4.01QAQKK745 pKa = 10.33LDD747 pKa = 3.35HH748 pKa = 6.09MAKK751 pKa = 10.28FFTFAPLSMHH761 pKa = 6.38KK762 pKa = 10.49KK763 pKa = 9.64IDD765 pKa = 3.54IEE767 pKa = 4.35SLSDD771 pKa = 3.69LLKK774 pKa = 10.46ISYY777 pKa = 9.74GPKK780 pKa = 10.1SLFLGKK786 pKa = 10.06DD787 pKa = 3.04DD788 pKa = 5.68WILKK792 pKa = 10.51DD793 pKa = 3.67PLTKK797 pKa = 10.2KK798 pKa = 10.43VLSKK802 pKa = 10.77KK803 pKa = 9.05VLSRR807 pKa = 11.84QRR809 pKa = 11.84VSKK812 pKa = 10.84KK813 pKa = 10.17DD814 pKa = 3.27VQNAIRR820 pKa = 11.84VSTFRR825 pKa = 11.84MAVMDD830 pKa = 3.59QYY832 pKa = 11.71LRR834 pKa = 11.84SNKK837 pKa = 9.76
Molecular weight: 95.44 kDa
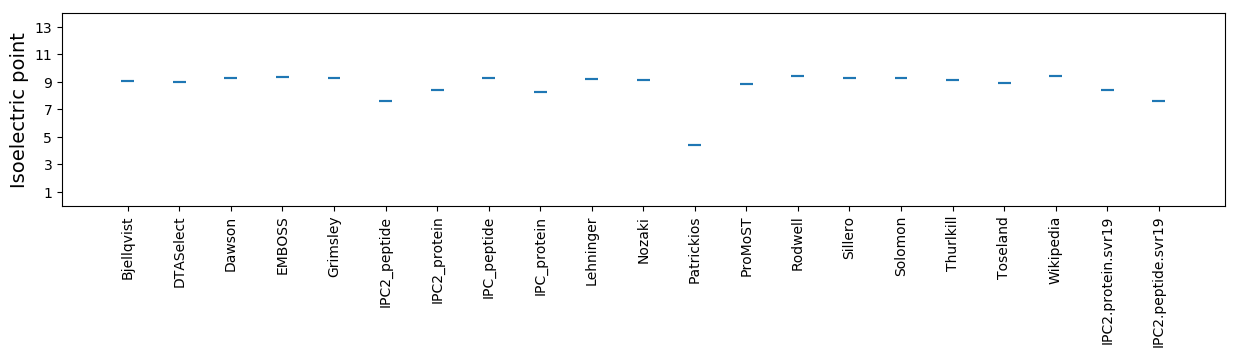
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIS0|A0A1L3KIS0_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 9 OX=1923509 PE=4 SV=1
MM1 pKa = 7.45ICILLMIMHH10 pKa = 7.26ILICVSRR17 pKa = 11.84SVQKK21 pKa = 10.85HH22 pKa = 4.95FFWSKK27 pKa = 9.73ANSWSAALKK36 pKa = 10.85NGDD39 pKa = 3.12GKK41 pKa = 11.12APNFDD46 pKa = 3.13RR47 pKa = 11.84KK48 pKa = 9.62MNQIVLSKK56 pKa = 9.58TYY58 pKa = 10.1KK59 pKa = 10.26ILMKK63 pKa = 10.45LSMGGVVNGILKK75 pKa = 10.36DD76 pKa = 3.1IYY78 pKa = 10.18TSHH81 pKa = 6.14NKK83 pKa = 9.92KK84 pKa = 10.28LEE86 pKa = 4.26KK87 pKa = 10.6YY88 pKa = 9.28SANAVIQQFKK98 pKa = 10.93EE99 pKa = 3.73DD100 pKa = 3.98FLFVKK105 pKa = 10.52NYY107 pKa = 10.26FLMGTVSEE115 pKa = 5.01FDD117 pKa = 4.17QKK119 pKa = 11.1LQWRR123 pKa = 11.84RR124 pKa = 11.84LSDD127 pKa = 3.19VGTFSLPPYY136 pKa = 9.88LSPIYY141 pKa = 10.26HH142 pKa = 7.27RR143 pKa = 11.84LGEE146 pKa = 4.21LNSWDD151 pKa = 3.72KK152 pKa = 11.59VLLLTLLRR160 pKa = 11.84SYY162 pKa = 11.29KK163 pKa = 10.75HH164 pKa = 5.75MDD166 pKa = 2.69IPGPHH171 pKa = 7.09KK172 pKa = 10.95LEE174 pKa = 4.64TIYY177 pKa = 11.21TMMGYY182 pKa = 7.68QTYY185 pKa = 9.64INTYY189 pKa = 7.83KK190 pKa = 10.4VCMSMSCVYY199 pKa = 10.71KK200 pKa = 10.34WMGMPKK206 pKa = 9.99LQPSDD211 pKa = 3.17TGDD214 pKa = 3.35YY215 pKa = 11.01SKK217 pKa = 11.04ISIAKK222 pKa = 9.68GLTPGTNTGSLLHH235 pKa = 6.61AVGPSGNTLVSRR247 pKa = 11.84LVDD250 pKa = 4.02LMNVYY255 pKa = 9.86QKK257 pKa = 11.12PMYY260 pKa = 10.56AGLLDD265 pKa = 4.73LFAKK269 pKa = 10.31FYY271 pKa = 11.3SDD273 pKa = 3.34FTKK276 pKa = 10.57VGKK279 pKa = 10.7GPGITNVSKK288 pKa = 10.95LFQHH292 pKa = 6.55FSKK295 pKa = 10.7PVLDD299 pKa = 4.29EE300 pKa = 4.09TLSMAKK306 pKa = 9.62EE307 pKa = 3.83NSKK310 pKa = 10.4IKK312 pKa = 10.36PKK314 pKa = 10.67VDD316 pKa = 3.18MKK318 pKa = 10.72ILNRR322 pKa = 11.84LSYY325 pKa = 9.47FTDD328 pKa = 4.29GAGKK332 pKa = 8.65WRR334 pKa = 11.84YY335 pKa = 8.9IAISDD340 pKa = 4.74WITQSILSPMHH351 pKa = 6.46NLMFDD356 pKa = 3.58ALKK359 pKa = 11.1SMSTDD364 pKa = 3.23CTFDD368 pKa = 3.3QNKK371 pKa = 8.84IRR373 pKa = 11.84AKK375 pKa = 7.85YY376 pKa = 9.79QEE378 pKa = 4.88MYY380 pKa = 10.46SQGKK384 pKa = 8.53GAYY387 pKa = 9.8CLDD390 pKa = 3.97LSAATDD396 pKa = 3.9RR397 pKa = 11.84LPLPLQAFLMFLLTKK412 pKa = 10.73NFGLSLSWMLIMMYY426 pKa = 10.6NPFHH430 pKa = 6.46LTGDD434 pKa = 3.81MEE436 pKa = 4.74KK437 pKa = 10.62KK438 pKa = 10.54FYY440 pKa = 10.34YY441 pKa = 9.05QTGQGMGIYY450 pKa = 9.4TSWAMLALTNHH461 pKa = 7.03AIVRR465 pKa = 11.84LSGKK469 pKa = 10.33VYY471 pKa = 9.94GHH473 pKa = 6.61SDD475 pKa = 3.18FSDD478 pKa = 3.76YY479 pKa = 11.3LVLGDD484 pKa = 5.29DD485 pKa = 3.53VVIFNKK491 pKa = 9.92EE492 pKa = 3.8VADD495 pKa = 4.7GYY497 pKa = 9.32LTVMSKK503 pKa = 10.87LGVDD507 pKa = 3.65ISQPKK512 pKa = 10.43SIFPKK517 pKa = 9.9EE518 pKa = 3.98NKK520 pKa = 9.57FGCEE524 pKa = 3.32FASRR528 pKa = 11.84LFVEE532 pKa = 4.89GQEE535 pKa = 4.31LSPLPVGLLIKK546 pKa = 10.44DD547 pKa = 3.8PNVDD551 pKa = 3.61NILNFWTTVHH561 pKa = 6.1WKK563 pKa = 8.19STEE566 pKa = 4.01LVEE569 pKa = 4.42TQYY572 pKa = 11.35QDD574 pKa = 4.09SLTTPDD580 pKa = 4.2LSSAVPLSGSEE591 pKa = 4.02DD592 pKa = 3.63LKK594 pKa = 10.35TIWGFMSVYY603 pKa = 10.09NYY605 pKa = 10.95YY606 pKa = 8.72YY607 pKa = 7.58TQHH610 pKa = 5.96MKK612 pKa = 10.43FSEE615 pKa = 4.21NEE617 pKa = 4.19VPSGAWMSLPEE628 pKa = 4.56ASPLHH633 pKa = 5.88TLLEE637 pKa = 4.81AIPLKK642 pKa = 10.98SMEE645 pKa = 4.32EE646 pKa = 4.16VEE648 pKa = 4.33STMEE652 pKa = 4.3KK653 pKa = 9.22ILYY656 pKa = 9.9RR657 pKa = 11.84RR658 pKa = 11.84FSQALKK664 pKa = 10.35TMTKK668 pKa = 9.89HH669 pKa = 6.58SKK671 pKa = 9.18TFDD674 pKa = 3.62SIKK677 pKa = 11.22DD678 pKa = 3.58MLVPLYY684 pKa = 10.85GSLGDD689 pKa = 3.74LEE691 pKa = 5.3SRR693 pKa = 11.84FPHH696 pKa = 6.46ILSMYY701 pKa = 9.55LFFMSPFYY709 pKa = 11.05HH710 pKa = 6.99CIDD713 pKa = 3.87KK714 pKa = 10.79FSEE717 pKa = 4.33LNLDD721 pKa = 3.9PQSEE725 pKa = 4.48MDD727 pKa = 3.31SHH729 pKa = 6.65MIAAGSGVLVMDD741 pKa = 4.01QAQKK745 pKa = 10.33LDD747 pKa = 3.35HH748 pKa = 6.09MAKK751 pKa = 10.28FFTFAPLSMHH761 pKa = 6.38KK762 pKa = 10.49KK763 pKa = 9.64IDD765 pKa = 3.54IEE767 pKa = 4.35SLSDD771 pKa = 3.69LLKK774 pKa = 10.46ISYY777 pKa = 9.74GPKK780 pKa = 10.1SLFLGKK786 pKa = 10.06DD787 pKa = 3.04DD788 pKa = 5.68WILKK792 pKa = 10.51DD793 pKa = 3.67PLTKK797 pKa = 10.2KK798 pKa = 10.43VLSKK802 pKa = 10.77KK803 pKa = 9.05VLSRR807 pKa = 11.84QRR809 pKa = 11.84VSKK812 pKa = 10.84KK813 pKa = 10.17DD814 pKa = 3.27VQNAIRR820 pKa = 11.84VSTFRR825 pKa = 11.84MAVMDD830 pKa = 3.59QYY832 pKa = 11.71LRR834 pKa = 11.84SNKK837 pKa = 9.76
MM1 pKa = 7.45ICILLMIMHH10 pKa = 7.26ILICVSRR17 pKa = 11.84SVQKK21 pKa = 10.85HH22 pKa = 4.95FFWSKK27 pKa = 9.73ANSWSAALKK36 pKa = 10.85NGDD39 pKa = 3.12GKK41 pKa = 11.12APNFDD46 pKa = 3.13RR47 pKa = 11.84KK48 pKa = 9.62MNQIVLSKK56 pKa = 9.58TYY58 pKa = 10.1KK59 pKa = 10.26ILMKK63 pKa = 10.45LSMGGVVNGILKK75 pKa = 10.36DD76 pKa = 3.1IYY78 pKa = 10.18TSHH81 pKa = 6.14NKK83 pKa = 9.92KK84 pKa = 10.28LEE86 pKa = 4.26KK87 pKa = 10.6YY88 pKa = 9.28SANAVIQQFKK98 pKa = 10.93EE99 pKa = 3.73DD100 pKa = 3.98FLFVKK105 pKa = 10.52NYY107 pKa = 10.26FLMGTVSEE115 pKa = 5.01FDD117 pKa = 4.17QKK119 pKa = 11.1LQWRR123 pKa = 11.84RR124 pKa = 11.84LSDD127 pKa = 3.19VGTFSLPPYY136 pKa = 9.88LSPIYY141 pKa = 10.26HH142 pKa = 7.27RR143 pKa = 11.84LGEE146 pKa = 4.21LNSWDD151 pKa = 3.72KK152 pKa = 11.59VLLLTLLRR160 pKa = 11.84SYY162 pKa = 11.29KK163 pKa = 10.75HH164 pKa = 5.75MDD166 pKa = 2.69IPGPHH171 pKa = 7.09KK172 pKa = 10.95LEE174 pKa = 4.64TIYY177 pKa = 11.21TMMGYY182 pKa = 7.68QTYY185 pKa = 9.64INTYY189 pKa = 7.83KK190 pKa = 10.4VCMSMSCVYY199 pKa = 10.71KK200 pKa = 10.34WMGMPKK206 pKa = 9.99LQPSDD211 pKa = 3.17TGDD214 pKa = 3.35YY215 pKa = 11.01SKK217 pKa = 11.04ISIAKK222 pKa = 9.68GLTPGTNTGSLLHH235 pKa = 6.61AVGPSGNTLVSRR247 pKa = 11.84LVDD250 pKa = 4.02LMNVYY255 pKa = 9.86QKK257 pKa = 11.12PMYY260 pKa = 10.56AGLLDD265 pKa = 4.73LFAKK269 pKa = 10.31FYY271 pKa = 11.3SDD273 pKa = 3.34FTKK276 pKa = 10.57VGKK279 pKa = 10.7GPGITNVSKK288 pKa = 10.95LFQHH292 pKa = 6.55FSKK295 pKa = 10.7PVLDD299 pKa = 4.29EE300 pKa = 4.09TLSMAKK306 pKa = 9.62EE307 pKa = 3.83NSKK310 pKa = 10.4IKK312 pKa = 10.36PKK314 pKa = 10.67VDD316 pKa = 3.18MKK318 pKa = 10.72ILNRR322 pKa = 11.84LSYY325 pKa = 9.47FTDD328 pKa = 4.29GAGKK332 pKa = 8.65WRR334 pKa = 11.84YY335 pKa = 8.9IAISDD340 pKa = 4.74WITQSILSPMHH351 pKa = 6.46NLMFDD356 pKa = 3.58ALKK359 pKa = 11.1SMSTDD364 pKa = 3.23CTFDD368 pKa = 3.3QNKK371 pKa = 8.84IRR373 pKa = 11.84AKK375 pKa = 7.85YY376 pKa = 9.79QEE378 pKa = 4.88MYY380 pKa = 10.46SQGKK384 pKa = 8.53GAYY387 pKa = 9.8CLDD390 pKa = 3.97LSAATDD396 pKa = 3.9RR397 pKa = 11.84LPLPLQAFLMFLLTKK412 pKa = 10.73NFGLSLSWMLIMMYY426 pKa = 10.6NPFHH430 pKa = 6.46LTGDD434 pKa = 3.81MEE436 pKa = 4.74KK437 pKa = 10.62KK438 pKa = 10.54FYY440 pKa = 10.34YY441 pKa = 9.05QTGQGMGIYY450 pKa = 9.4TSWAMLALTNHH461 pKa = 7.03AIVRR465 pKa = 11.84LSGKK469 pKa = 10.33VYY471 pKa = 9.94GHH473 pKa = 6.61SDD475 pKa = 3.18FSDD478 pKa = 3.76YY479 pKa = 11.3LVLGDD484 pKa = 5.29DD485 pKa = 3.53VVIFNKK491 pKa = 9.92EE492 pKa = 3.8VADD495 pKa = 4.7GYY497 pKa = 9.32LTVMSKK503 pKa = 10.87LGVDD507 pKa = 3.65ISQPKK512 pKa = 10.43SIFPKK517 pKa = 9.9EE518 pKa = 3.98NKK520 pKa = 9.57FGCEE524 pKa = 3.32FASRR528 pKa = 11.84LFVEE532 pKa = 4.89GQEE535 pKa = 4.31LSPLPVGLLIKK546 pKa = 10.44DD547 pKa = 3.8PNVDD551 pKa = 3.61NILNFWTTVHH561 pKa = 6.1WKK563 pKa = 8.19STEE566 pKa = 4.01LVEE569 pKa = 4.42TQYY572 pKa = 11.35QDD574 pKa = 4.09SLTTPDD580 pKa = 4.2LSSAVPLSGSEE591 pKa = 4.02DD592 pKa = 3.63LKK594 pKa = 10.35TIWGFMSVYY603 pKa = 10.09NYY605 pKa = 10.95YY606 pKa = 8.72YY607 pKa = 7.58TQHH610 pKa = 5.96MKK612 pKa = 10.43FSEE615 pKa = 4.21NEE617 pKa = 4.19VPSGAWMSLPEE628 pKa = 4.56ASPLHH633 pKa = 5.88TLLEE637 pKa = 4.81AIPLKK642 pKa = 10.98SMEE645 pKa = 4.32EE646 pKa = 4.16VEE648 pKa = 4.33STMEE652 pKa = 4.3KK653 pKa = 9.22ILYY656 pKa = 9.9RR657 pKa = 11.84RR658 pKa = 11.84FSQALKK664 pKa = 10.35TMTKK668 pKa = 9.89HH669 pKa = 6.58SKK671 pKa = 9.18TFDD674 pKa = 3.62SIKK677 pKa = 11.22DD678 pKa = 3.58MLVPLYY684 pKa = 10.85GSLGDD689 pKa = 3.74LEE691 pKa = 5.3SRR693 pKa = 11.84FPHH696 pKa = 6.46ILSMYY701 pKa = 9.55LFFMSPFYY709 pKa = 11.05HH710 pKa = 6.99CIDD713 pKa = 3.87KK714 pKa = 10.79FSEE717 pKa = 4.33LNLDD721 pKa = 3.9PQSEE725 pKa = 4.48MDD727 pKa = 3.31SHH729 pKa = 6.65MIAAGSGVLVMDD741 pKa = 4.01QAQKK745 pKa = 10.33LDD747 pKa = 3.35HH748 pKa = 6.09MAKK751 pKa = 10.28FFTFAPLSMHH761 pKa = 6.38KK762 pKa = 10.49KK763 pKa = 9.64IDD765 pKa = 3.54IEE767 pKa = 4.35SLSDD771 pKa = 3.69LLKK774 pKa = 10.46ISYY777 pKa = 9.74GPKK780 pKa = 10.1SLFLGKK786 pKa = 10.06DD787 pKa = 3.04DD788 pKa = 5.68WILKK792 pKa = 10.51DD793 pKa = 3.67PLTKK797 pKa = 10.2KK798 pKa = 10.43VLSKK802 pKa = 10.77KK803 pKa = 9.05VLSRR807 pKa = 11.84QRR809 pKa = 11.84VSKK812 pKa = 10.84KK813 pKa = 10.17DD814 pKa = 3.27VQNAIRR820 pKa = 11.84VSTFRR825 pKa = 11.84MAVMDD830 pKa = 3.59QYY832 pKa = 11.71LRR834 pKa = 11.84SNKK837 pKa = 9.76
Molecular weight: 95.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
837 |
837 |
837 |
837.0 |
95.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.301 ± 0.0 | 0.956 ± 0.0 |
5.854 ± 0.0 | 3.465 ± 0.0 |
5.137 ± 0.0 | 5.615 ± 0.0 |
2.509 ± 0.0 | 5.257 ± 0.0 |
8.841 ± 0.0 | 11.828 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
5.496 ± 0.0 | 3.704 ± 0.0 |
4.301 ± 0.0 | 3.465 ± 0.0 |
2.509 ± 0.0 | 9.677 ± 0.0 |
5.257 ± 0.0 | 5.496 ± 0.0 |
1.673 ± 0.0 | 4.659 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
