
Cryphonectria nitschkei chrysovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Chrysoviridae; Alphachrysovirus
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
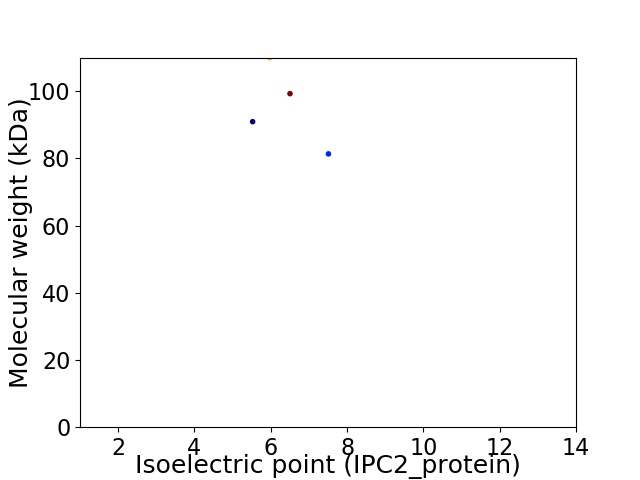
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D6QX90|D6QX90_9VIRU Putative replication associated protein OS=Cryphonectria nitschkei chrysovirus 1 OX=399394 PE=4 SV=1
MM1 pKa = 7.24SAYY4 pKa = 9.42KK5 pKa = 10.1RR6 pKa = 11.84VMDD9 pKa = 4.19YY10 pKa = 10.74MSGAPDD16 pKa = 3.42SANHH20 pKa = 5.52IKK22 pKa = 10.42LWEE25 pKa = 4.18KK26 pKa = 10.88AQMDD30 pKa = 4.02RR31 pKa = 11.84MRR33 pKa = 11.84DD34 pKa = 3.38QVDD37 pKa = 3.07ALEE40 pKa = 4.32LEE42 pKa = 4.19KK43 pKa = 11.1RR44 pKa = 11.84MLGPAEE50 pKa = 4.57RR51 pKa = 11.84DD52 pKa = 2.97LMAAFHH58 pKa = 6.29CQPKK62 pKa = 8.54FTRR65 pKa = 11.84KK66 pKa = 9.38GMNEE70 pKa = 3.7GLTIGVNVDD79 pKa = 3.24CRR81 pKa = 11.84TSSGDD86 pKa = 3.11KK87 pKa = 10.11VSRR90 pKa = 11.84IKK92 pKa = 11.1NVVVDD97 pKa = 5.03FNGGEE102 pKa = 4.23CWEE105 pKa = 4.3IAHH108 pKa = 6.91AAAHH112 pKa = 5.79VDD114 pKa = 3.62YY115 pKa = 10.04VTRR118 pKa = 11.84DD119 pKa = 3.32PAFYY123 pKa = 10.17TLDD126 pKa = 3.03TTLRR130 pKa = 11.84DD131 pKa = 4.24KK132 pKa = 10.82IISQCPGMSAAQLYY146 pKa = 6.91EE147 pKa = 3.83LRR149 pKa = 11.84VDD151 pKa = 3.84GVKK154 pKa = 10.65DD155 pKa = 3.79SLSLPVDD162 pKa = 3.38MRR164 pKa = 11.84HH165 pKa = 5.87MYY167 pKa = 9.73MKK169 pKa = 10.1MMLMLWDD176 pKa = 3.6YY177 pKa = 10.5TLAATSKK184 pKa = 9.88VEE186 pKa = 4.05GMEE189 pKa = 3.97MAEE192 pKa = 4.59DD193 pKa = 3.5IVDD196 pKa = 4.81FSHH199 pKa = 7.65PSLPADD205 pKa = 2.98ARR207 pKa = 11.84MATVANYY214 pKa = 10.08RR215 pKa = 11.84VVVDD219 pKa = 3.62AEE221 pKa = 4.3GMSRR225 pKa = 11.84RR226 pKa = 11.84EE227 pKa = 3.96LGLLSMAGQQYY238 pKa = 9.8PSVWYY243 pKa = 10.27AGDD246 pKa = 3.87NIYY249 pKa = 9.43TKK251 pKa = 10.66CHH253 pKa = 5.72LAEE256 pKa = 5.25DD257 pKa = 3.93SLAIVSDD264 pKa = 3.69GKK266 pKa = 10.61IDD268 pKa = 3.73MDD270 pKa = 3.99TSGMWGSPDD279 pKa = 3.35EE280 pKa = 4.85LYY282 pKa = 11.1SIIWSIACKK291 pKa = 9.83TSSTKK296 pKa = 10.53CLAEE300 pKa = 4.98AITLLRR306 pKa = 11.84GKK308 pKa = 8.62CQHH311 pKa = 6.28MADD314 pKa = 3.67FTKK317 pKa = 10.4RR318 pKa = 11.84AGPITVQSGVPLSVCKK334 pKa = 10.53VRR336 pKa = 11.84SVGAASASTILQSQPGYY353 pKa = 10.55FSTSLSLVTDD363 pKa = 4.06LLYY366 pKa = 11.28GKK368 pKa = 8.9MFEE371 pKa = 4.18ISATSVVEE379 pKa = 3.98QVGGLGDD386 pKa = 3.79RR387 pKa = 11.84TCGEE391 pKa = 4.27VPSTDD396 pKa = 2.75RR397 pKa = 11.84MYY399 pKa = 11.33NGLLRR404 pKa = 11.84DD405 pKa = 2.99IGLRR409 pKa = 11.84HH410 pKa = 6.61EE411 pKa = 5.34DD412 pKa = 3.37GSINAVLKK420 pKa = 9.88NWCSLTGSAYY430 pKa = 8.73TFGYY434 pKa = 10.06GGAIKK439 pKa = 10.3DD440 pKa = 3.83YY441 pKa = 10.54VIKK444 pKa = 9.83LTEE447 pKa = 4.04LMRR450 pKa = 11.84GGWDD454 pKa = 3.26VEE456 pKa = 4.22LPQLGTLIPFMDD468 pKa = 4.37TKK470 pKa = 9.97DD471 pKa = 3.64TCWGYY476 pKa = 11.27SRR478 pKa = 11.84GYY480 pKa = 8.78PGKK483 pKa = 9.54PDD485 pKa = 4.35VFDD488 pKa = 4.95KK489 pKa = 11.03DD490 pKa = 3.53QTMKK494 pKa = 10.8ASEE497 pKa = 4.31SLSVCAWLMGLRR509 pKa = 11.84ATRR512 pKa = 11.84PKK514 pKa = 10.47VGHH517 pKa = 6.01NRR519 pKa = 11.84SKK521 pKa = 10.87KK522 pKa = 8.43EE523 pKa = 3.9SRR525 pKa = 11.84HH526 pKa = 3.76QTPEE530 pKa = 3.61EE531 pKa = 3.93RR532 pKa = 11.84EE533 pKa = 3.9LAVEE537 pKa = 4.51SKK539 pKa = 11.07GKK541 pKa = 8.98MVLGSAQLWITDD553 pKa = 3.69GLGGRR558 pKa = 11.84EE559 pKa = 3.95DD560 pKa = 4.49DD561 pKa = 3.99YY562 pKa = 11.98EE563 pKa = 4.39EE564 pKa = 4.93GSGVLVKK571 pKa = 10.77SSFPSVKK578 pKa = 10.22CQLLYY583 pKa = 10.78DD584 pKa = 3.98EE585 pKa = 6.1RR586 pKa = 11.84DD587 pKa = 3.89QEE589 pKa = 4.14WHH591 pKa = 6.52IPTSRR596 pKa = 11.84EE597 pKa = 3.1RR598 pKa = 11.84DD599 pKa = 3.5YY600 pKa = 11.38TSMVRR605 pKa = 11.84RR606 pKa = 11.84TTMGEE611 pKa = 4.03HH612 pKa = 5.58EE613 pKa = 4.67VEE615 pKa = 4.26VAPVIVGPAPKK626 pKa = 10.14SPEE629 pKa = 4.48DD630 pKa = 3.24IFSGVRR636 pKa = 11.84AAVKK640 pKa = 9.2EE641 pKa = 4.22RR642 pKa = 11.84PVRR645 pKa = 11.84PYY647 pKa = 10.62KK648 pKa = 10.32EE649 pKa = 3.86PRR651 pKa = 11.84PAVGMTGVKK660 pKa = 9.83FVPRR664 pKa = 11.84YY665 pKa = 9.27VLDD668 pKa = 4.1GKK670 pKa = 10.42VVEE673 pKa = 4.98DD674 pKa = 5.05PIDD677 pKa = 3.55VRR679 pKa = 11.84TKK681 pKa = 10.47RR682 pKa = 11.84PATGEE687 pKa = 4.06MRR689 pKa = 11.84VKK691 pKa = 10.78AIDD694 pKa = 3.42VPGDD698 pKa = 3.96GKK700 pKa = 11.01CGLHH704 pKa = 6.86AVVQDD709 pKa = 4.09LQSHH713 pKa = 6.38GLLTQRR719 pKa = 11.84DD720 pKa = 3.89GGRR723 pKa = 11.84VLRR726 pKa = 11.84LLDD729 pKa = 3.75KK730 pKa = 10.23STNAPTFHH738 pKa = 7.26SPEE741 pKa = 5.51DD742 pKa = 3.34IAGAVLKK749 pKa = 10.77LGLGLDD755 pKa = 3.86VVAGGIVHH763 pKa = 7.12SYY765 pKa = 10.23GNNDD769 pKa = 3.04GHH771 pKa = 6.46RR772 pKa = 11.84VLMQLKK778 pKa = 8.77DD779 pKa = 3.49HH780 pKa = 7.04HH781 pKa = 6.97YY782 pKa = 8.64MPIVEE787 pKa = 4.41HH788 pKa = 6.91EE789 pKa = 4.37EE790 pKa = 4.58GEE792 pKa = 4.52TLNITSVQEE801 pKa = 3.93QVGTDD806 pKa = 3.21DD807 pKa = 4.99EE808 pKa = 4.91YY809 pKa = 10.95IKK811 pKa = 10.86RR812 pKa = 11.84LGEE815 pKa = 4.07FEE817 pKa = 4.79RR818 pKa = 11.84MLAAATT824 pKa = 3.76
MM1 pKa = 7.24SAYY4 pKa = 9.42KK5 pKa = 10.1RR6 pKa = 11.84VMDD9 pKa = 4.19YY10 pKa = 10.74MSGAPDD16 pKa = 3.42SANHH20 pKa = 5.52IKK22 pKa = 10.42LWEE25 pKa = 4.18KK26 pKa = 10.88AQMDD30 pKa = 4.02RR31 pKa = 11.84MRR33 pKa = 11.84DD34 pKa = 3.38QVDD37 pKa = 3.07ALEE40 pKa = 4.32LEE42 pKa = 4.19KK43 pKa = 11.1RR44 pKa = 11.84MLGPAEE50 pKa = 4.57RR51 pKa = 11.84DD52 pKa = 2.97LMAAFHH58 pKa = 6.29CQPKK62 pKa = 8.54FTRR65 pKa = 11.84KK66 pKa = 9.38GMNEE70 pKa = 3.7GLTIGVNVDD79 pKa = 3.24CRR81 pKa = 11.84TSSGDD86 pKa = 3.11KK87 pKa = 10.11VSRR90 pKa = 11.84IKK92 pKa = 11.1NVVVDD97 pKa = 5.03FNGGEE102 pKa = 4.23CWEE105 pKa = 4.3IAHH108 pKa = 6.91AAAHH112 pKa = 5.79VDD114 pKa = 3.62YY115 pKa = 10.04VTRR118 pKa = 11.84DD119 pKa = 3.32PAFYY123 pKa = 10.17TLDD126 pKa = 3.03TTLRR130 pKa = 11.84DD131 pKa = 4.24KK132 pKa = 10.82IISQCPGMSAAQLYY146 pKa = 6.91EE147 pKa = 3.83LRR149 pKa = 11.84VDD151 pKa = 3.84GVKK154 pKa = 10.65DD155 pKa = 3.79SLSLPVDD162 pKa = 3.38MRR164 pKa = 11.84HH165 pKa = 5.87MYY167 pKa = 9.73MKK169 pKa = 10.1MMLMLWDD176 pKa = 3.6YY177 pKa = 10.5TLAATSKK184 pKa = 9.88VEE186 pKa = 4.05GMEE189 pKa = 3.97MAEE192 pKa = 4.59DD193 pKa = 3.5IVDD196 pKa = 4.81FSHH199 pKa = 7.65PSLPADD205 pKa = 2.98ARR207 pKa = 11.84MATVANYY214 pKa = 10.08RR215 pKa = 11.84VVVDD219 pKa = 3.62AEE221 pKa = 4.3GMSRR225 pKa = 11.84RR226 pKa = 11.84EE227 pKa = 3.96LGLLSMAGQQYY238 pKa = 9.8PSVWYY243 pKa = 10.27AGDD246 pKa = 3.87NIYY249 pKa = 9.43TKK251 pKa = 10.66CHH253 pKa = 5.72LAEE256 pKa = 5.25DD257 pKa = 3.93SLAIVSDD264 pKa = 3.69GKK266 pKa = 10.61IDD268 pKa = 3.73MDD270 pKa = 3.99TSGMWGSPDD279 pKa = 3.35EE280 pKa = 4.85LYY282 pKa = 11.1SIIWSIACKK291 pKa = 9.83TSSTKK296 pKa = 10.53CLAEE300 pKa = 4.98AITLLRR306 pKa = 11.84GKK308 pKa = 8.62CQHH311 pKa = 6.28MADD314 pKa = 3.67FTKK317 pKa = 10.4RR318 pKa = 11.84AGPITVQSGVPLSVCKK334 pKa = 10.53VRR336 pKa = 11.84SVGAASASTILQSQPGYY353 pKa = 10.55FSTSLSLVTDD363 pKa = 4.06LLYY366 pKa = 11.28GKK368 pKa = 8.9MFEE371 pKa = 4.18ISATSVVEE379 pKa = 3.98QVGGLGDD386 pKa = 3.79RR387 pKa = 11.84TCGEE391 pKa = 4.27VPSTDD396 pKa = 2.75RR397 pKa = 11.84MYY399 pKa = 11.33NGLLRR404 pKa = 11.84DD405 pKa = 2.99IGLRR409 pKa = 11.84HH410 pKa = 6.61EE411 pKa = 5.34DD412 pKa = 3.37GSINAVLKK420 pKa = 9.88NWCSLTGSAYY430 pKa = 8.73TFGYY434 pKa = 10.06GGAIKK439 pKa = 10.3DD440 pKa = 3.83YY441 pKa = 10.54VIKK444 pKa = 9.83LTEE447 pKa = 4.04LMRR450 pKa = 11.84GGWDD454 pKa = 3.26VEE456 pKa = 4.22LPQLGTLIPFMDD468 pKa = 4.37TKK470 pKa = 9.97DD471 pKa = 3.64TCWGYY476 pKa = 11.27SRR478 pKa = 11.84GYY480 pKa = 8.78PGKK483 pKa = 9.54PDD485 pKa = 4.35VFDD488 pKa = 4.95KK489 pKa = 11.03DD490 pKa = 3.53QTMKK494 pKa = 10.8ASEE497 pKa = 4.31SLSVCAWLMGLRR509 pKa = 11.84ATRR512 pKa = 11.84PKK514 pKa = 10.47VGHH517 pKa = 6.01NRR519 pKa = 11.84SKK521 pKa = 10.87KK522 pKa = 8.43EE523 pKa = 3.9SRR525 pKa = 11.84HH526 pKa = 3.76QTPEE530 pKa = 3.61EE531 pKa = 3.93RR532 pKa = 11.84EE533 pKa = 3.9LAVEE537 pKa = 4.51SKK539 pKa = 11.07GKK541 pKa = 8.98MVLGSAQLWITDD553 pKa = 3.69GLGGRR558 pKa = 11.84EE559 pKa = 3.95DD560 pKa = 4.49DD561 pKa = 3.99YY562 pKa = 11.98EE563 pKa = 4.39EE564 pKa = 4.93GSGVLVKK571 pKa = 10.77SSFPSVKK578 pKa = 10.22CQLLYY583 pKa = 10.78DD584 pKa = 3.98EE585 pKa = 6.1RR586 pKa = 11.84DD587 pKa = 3.89QEE589 pKa = 4.14WHH591 pKa = 6.52IPTSRR596 pKa = 11.84EE597 pKa = 3.1RR598 pKa = 11.84DD599 pKa = 3.5YY600 pKa = 11.38TSMVRR605 pKa = 11.84RR606 pKa = 11.84TTMGEE611 pKa = 4.03HH612 pKa = 5.58EE613 pKa = 4.67VEE615 pKa = 4.26VAPVIVGPAPKK626 pKa = 10.14SPEE629 pKa = 4.48DD630 pKa = 3.24IFSGVRR636 pKa = 11.84AAVKK640 pKa = 9.2EE641 pKa = 4.22RR642 pKa = 11.84PVRR645 pKa = 11.84PYY647 pKa = 10.62KK648 pKa = 10.32EE649 pKa = 3.86PRR651 pKa = 11.84PAVGMTGVKK660 pKa = 9.83FVPRR664 pKa = 11.84YY665 pKa = 9.27VLDD668 pKa = 4.1GKK670 pKa = 10.42VVEE673 pKa = 4.98DD674 pKa = 5.05PIDD677 pKa = 3.55VRR679 pKa = 11.84TKK681 pKa = 10.47RR682 pKa = 11.84PATGEE687 pKa = 4.06MRR689 pKa = 11.84VKK691 pKa = 10.78AIDD694 pKa = 3.42VPGDD698 pKa = 3.96GKK700 pKa = 11.01CGLHH704 pKa = 6.86AVVQDD709 pKa = 4.09LQSHH713 pKa = 6.38GLLTQRR719 pKa = 11.84DD720 pKa = 3.89GGRR723 pKa = 11.84VLRR726 pKa = 11.84LLDD729 pKa = 3.75KK730 pKa = 10.23STNAPTFHH738 pKa = 7.26SPEE741 pKa = 5.51DD742 pKa = 3.34IAGAVLKK749 pKa = 10.77LGLGLDD755 pKa = 3.86VVAGGIVHH763 pKa = 7.12SYY765 pKa = 10.23GNNDD769 pKa = 3.04GHH771 pKa = 6.46RR772 pKa = 11.84VLMQLKK778 pKa = 8.77DD779 pKa = 3.49HH780 pKa = 7.04HH781 pKa = 6.97YY782 pKa = 8.64MPIVEE787 pKa = 4.41HH788 pKa = 6.91EE789 pKa = 4.37EE790 pKa = 4.58GEE792 pKa = 4.52TLNITSVQEE801 pKa = 3.93QVGTDD806 pKa = 3.21DD807 pKa = 4.99EE808 pKa = 4.91YY809 pKa = 10.95IKK811 pKa = 10.86RR812 pKa = 11.84LGEE815 pKa = 4.07FEE817 pKa = 4.79RR818 pKa = 11.84MLAAATT824 pKa = 3.76
Molecular weight: 90.96 kDa
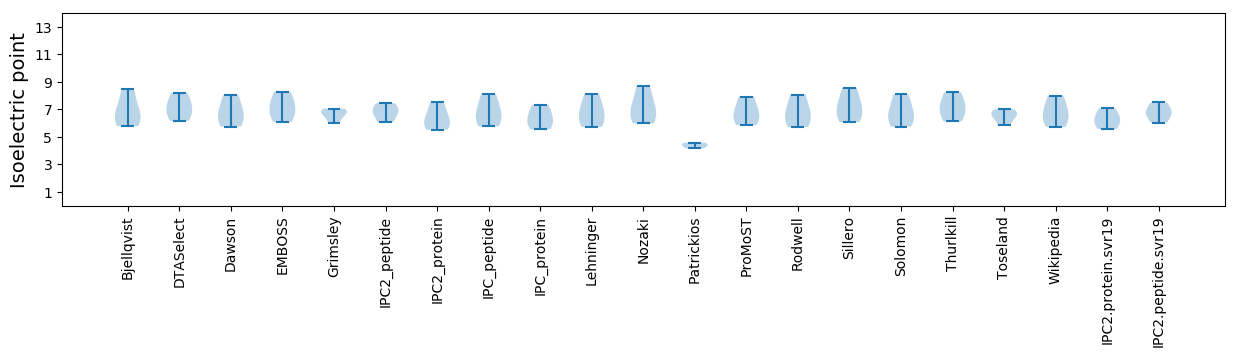
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D6QX90|D6QX90_9VIRU Putative replication associated protein OS=Cryphonectria nitschkei chrysovirus 1 OX=399394 PE=4 SV=1
MM1 pKa = 7.53SISPNSRR8 pKa = 11.84YY9 pKa = 10.31AGGGGHH15 pKa = 5.24GHH17 pKa = 6.52KK18 pKa = 10.64VGLAEE23 pKa = 3.9KK24 pKa = 10.18AARR27 pKa = 11.84RR28 pKa = 11.84LINRR32 pKa = 11.84GKK34 pKa = 8.69VTYY37 pKa = 10.01GPKK40 pKa = 10.44DD41 pKa = 3.75FEE43 pKa = 4.82VKK45 pKa = 10.67DD46 pKa = 3.58SDD48 pKa = 3.79RR49 pKa = 11.84NLIGVVIPSLLGKK62 pKa = 6.83TTLANQFGWLDD73 pKa = 3.58VDD75 pKa = 6.25DD76 pKa = 5.08LTTDD80 pKa = 3.54DD81 pKa = 5.46KK82 pKa = 11.61RR83 pKa = 11.84NQLFATFSQRR93 pKa = 11.84YY94 pKa = 9.09AEE96 pKa = 5.1DD97 pKa = 3.62GWCAAMRR104 pKa = 11.84PFNEE108 pKa = 4.08EE109 pKa = 3.7CEE111 pKa = 4.04ASIDD115 pKa = 3.44RR116 pKa = 11.84MYY118 pKa = 10.67FYY120 pKa = 11.0SQTVILAHH128 pKa = 6.07SVSSLAYY135 pKa = 9.98LGVIPSIRR143 pKa = 11.84IAPSDD148 pKa = 3.69DD149 pKa = 3.31FAAKK153 pKa = 10.25LLSLEE158 pKa = 4.48TGGSAAFAKK167 pKa = 10.73ANIEE171 pKa = 4.06AVRR174 pKa = 11.84EE175 pKa = 4.11EE176 pKa = 4.66ADD178 pKa = 3.38SSPLVTVSSYY188 pKa = 11.87DD189 pKa = 3.65EE190 pKa = 4.41LSATAVQLAIRR201 pKa = 11.84LGHH204 pKa = 7.03DD205 pKa = 3.58VDD207 pKa = 5.08CKK209 pKa = 10.97AVSQDD214 pKa = 3.67DD215 pKa = 5.64AIDD218 pKa = 3.53QYY220 pKa = 11.87EE221 pKa = 4.32CGLIEE226 pKa = 5.89RR227 pKa = 11.84DD228 pKa = 3.52DD229 pKa = 3.93ADD231 pKa = 3.9RR232 pKa = 11.84LVRR235 pKa = 11.84EE236 pKa = 4.2HH237 pKa = 6.95GGLHH241 pKa = 6.33RR242 pKa = 11.84GFGRR246 pKa = 11.84TRR248 pKa = 11.84SGWARR253 pKa = 11.84AVACIGCDD261 pKa = 3.27NKK263 pKa = 10.5PALSVTVDD271 pKa = 3.61KK272 pKa = 11.17EE273 pKa = 4.4VLDD276 pKa = 3.98KK277 pKa = 11.48ANCINRR283 pKa = 11.84AVVGRR288 pKa = 11.84ILTKK292 pKa = 10.28SNKK295 pKa = 8.77TSAHH299 pKa = 4.46VQALLTFVVAFGKK312 pKa = 10.56GQLLDD317 pKa = 4.02VISNLLTTPEE327 pKa = 4.23SDD329 pKa = 2.93WEE331 pKa = 4.0RR332 pKa = 11.84VMRR335 pKa = 11.84GISSLVKK342 pKa = 9.9TSNSYY347 pKa = 9.32MGYY350 pKa = 10.41DD351 pKa = 3.27LSDD354 pKa = 3.56DD355 pKa = 3.59EE356 pKa = 4.54RR357 pKa = 11.84RR358 pKa = 11.84VMSEE362 pKa = 3.42LWLLGHH368 pKa = 6.47QNRR371 pKa = 11.84TSMAALMRR379 pKa = 11.84EE380 pKa = 4.0WLLGCGGTYY389 pKa = 10.5KK390 pKa = 10.77SIPTSADD397 pKa = 2.36IRR399 pKa = 11.84KK400 pKa = 8.97AADD403 pKa = 3.39GAGLAGGRR411 pKa = 11.84TVRR414 pKa = 11.84QSLQQALKK422 pKa = 8.88MWAEE426 pKa = 4.33TWRR429 pKa = 11.84LSDD432 pKa = 3.61KK433 pKa = 11.24ASLEE437 pKa = 3.94ALQGVDD443 pKa = 3.88LAFGNASGTSWYY455 pKa = 9.29KK456 pKa = 9.51ACRR459 pKa = 11.84IIYY462 pKa = 7.73LTYY465 pKa = 10.01EE466 pKa = 3.78SDD468 pKa = 5.45EE469 pKa = 4.18IGEE472 pKa = 4.26WLSKK476 pKa = 10.28LVSVRR481 pKa = 11.84EE482 pKa = 3.84QDD484 pKa = 3.4VRR486 pKa = 11.84EE487 pKa = 4.12VDD489 pKa = 2.78WKK491 pKa = 10.98RR492 pKa = 11.84RR493 pKa = 11.84VEE495 pKa = 4.24SAVSSLLLNLLAGAIGGIEE514 pKa = 4.18VEE516 pKa = 4.28KK517 pKa = 10.68HH518 pKa = 4.83SCPVRR523 pKa = 11.84ASDD526 pKa = 4.95CDD528 pKa = 3.57ALSAVILMMGIRR540 pKa = 11.84QVEE543 pKa = 4.27RR544 pKa = 11.84DD545 pKa = 3.5TGGLTTWAAGRR556 pKa = 11.84IAEE559 pKa = 4.51TMTMMEE565 pKa = 4.61SKK567 pKa = 10.62VVTAIEE573 pKa = 4.16ASLIMSSGDD582 pKa = 3.25KK583 pKa = 10.82AHH585 pKa = 6.71EE586 pKa = 3.76LWANIWQRR594 pKa = 11.84GAIYY598 pKa = 10.14HH599 pKa = 5.63MPGMMWMARR608 pKa = 11.84HH609 pKa = 5.89KK610 pKa = 10.22FDD612 pKa = 4.03HH613 pKa = 6.43KK614 pKa = 10.6RR615 pKa = 11.84RR616 pKa = 11.84SDD618 pKa = 3.83GYY620 pKa = 9.51IKK622 pKa = 10.57AYY624 pKa = 9.9LARR627 pKa = 11.84LCSPKK632 pKa = 10.66SKK634 pKa = 10.42GGSGRR639 pKa = 11.84VSMTGSTFHH648 pKa = 7.53PEE650 pKa = 3.75QRR652 pKa = 11.84WSGKK656 pKa = 9.52HH657 pKa = 4.41KK658 pKa = 10.59QQGEE662 pKa = 3.85WRR664 pKa = 11.84RR665 pKa = 11.84ASGSTSLVEE674 pKa = 3.73LRR676 pKa = 11.84GIEE679 pKa = 4.09DD680 pKa = 3.82VPTISCGEE688 pKa = 4.18AIKK691 pKa = 9.46STKK694 pKa = 10.25AVALGLIGSCITQGLPAKK712 pKa = 9.5KK713 pKa = 9.12VCVVLGRR720 pKa = 11.84LLAKK724 pKa = 10.17RR725 pKa = 11.84RR726 pKa = 11.84QVLIGKK732 pKa = 7.59VEE734 pKa = 4.01KK735 pKa = 11.06VEE737 pKa = 4.48EE738 pKa = 4.51PEE740 pKa = 3.96QQLL743 pKa = 3.46
MM1 pKa = 7.53SISPNSRR8 pKa = 11.84YY9 pKa = 10.31AGGGGHH15 pKa = 5.24GHH17 pKa = 6.52KK18 pKa = 10.64VGLAEE23 pKa = 3.9KK24 pKa = 10.18AARR27 pKa = 11.84RR28 pKa = 11.84LINRR32 pKa = 11.84GKK34 pKa = 8.69VTYY37 pKa = 10.01GPKK40 pKa = 10.44DD41 pKa = 3.75FEE43 pKa = 4.82VKK45 pKa = 10.67DD46 pKa = 3.58SDD48 pKa = 3.79RR49 pKa = 11.84NLIGVVIPSLLGKK62 pKa = 6.83TTLANQFGWLDD73 pKa = 3.58VDD75 pKa = 6.25DD76 pKa = 5.08LTTDD80 pKa = 3.54DD81 pKa = 5.46KK82 pKa = 11.61RR83 pKa = 11.84NQLFATFSQRR93 pKa = 11.84YY94 pKa = 9.09AEE96 pKa = 5.1DD97 pKa = 3.62GWCAAMRR104 pKa = 11.84PFNEE108 pKa = 4.08EE109 pKa = 3.7CEE111 pKa = 4.04ASIDD115 pKa = 3.44RR116 pKa = 11.84MYY118 pKa = 10.67FYY120 pKa = 11.0SQTVILAHH128 pKa = 6.07SVSSLAYY135 pKa = 9.98LGVIPSIRR143 pKa = 11.84IAPSDD148 pKa = 3.69DD149 pKa = 3.31FAAKK153 pKa = 10.25LLSLEE158 pKa = 4.48TGGSAAFAKK167 pKa = 10.73ANIEE171 pKa = 4.06AVRR174 pKa = 11.84EE175 pKa = 4.11EE176 pKa = 4.66ADD178 pKa = 3.38SSPLVTVSSYY188 pKa = 11.87DD189 pKa = 3.65EE190 pKa = 4.41LSATAVQLAIRR201 pKa = 11.84LGHH204 pKa = 7.03DD205 pKa = 3.58VDD207 pKa = 5.08CKK209 pKa = 10.97AVSQDD214 pKa = 3.67DD215 pKa = 5.64AIDD218 pKa = 3.53QYY220 pKa = 11.87EE221 pKa = 4.32CGLIEE226 pKa = 5.89RR227 pKa = 11.84DD228 pKa = 3.52DD229 pKa = 3.93ADD231 pKa = 3.9RR232 pKa = 11.84LVRR235 pKa = 11.84EE236 pKa = 4.2HH237 pKa = 6.95GGLHH241 pKa = 6.33RR242 pKa = 11.84GFGRR246 pKa = 11.84TRR248 pKa = 11.84SGWARR253 pKa = 11.84AVACIGCDD261 pKa = 3.27NKK263 pKa = 10.5PALSVTVDD271 pKa = 3.61KK272 pKa = 11.17EE273 pKa = 4.4VLDD276 pKa = 3.98KK277 pKa = 11.48ANCINRR283 pKa = 11.84AVVGRR288 pKa = 11.84ILTKK292 pKa = 10.28SNKK295 pKa = 8.77TSAHH299 pKa = 4.46VQALLTFVVAFGKK312 pKa = 10.56GQLLDD317 pKa = 4.02VISNLLTTPEE327 pKa = 4.23SDD329 pKa = 2.93WEE331 pKa = 4.0RR332 pKa = 11.84VMRR335 pKa = 11.84GISSLVKK342 pKa = 9.9TSNSYY347 pKa = 9.32MGYY350 pKa = 10.41DD351 pKa = 3.27LSDD354 pKa = 3.56DD355 pKa = 3.59EE356 pKa = 4.54RR357 pKa = 11.84RR358 pKa = 11.84VMSEE362 pKa = 3.42LWLLGHH368 pKa = 6.47QNRR371 pKa = 11.84TSMAALMRR379 pKa = 11.84EE380 pKa = 4.0WLLGCGGTYY389 pKa = 10.5KK390 pKa = 10.77SIPTSADD397 pKa = 2.36IRR399 pKa = 11.84KK400 pKa = 8.97AADD403 pKa = 3.39GAGLAGGRR411 pKa = 11.84TVRR414 pKa = 11.84QSLQQALKK422 pKa = 8.88MWAEE426 pKa = 4.33TWRR429 pKa = 11.84LSDD432 pKa = 3.61KK433 pKa = 11.24ASLEE437 pKa = 3.94ALQGVDD443 pKa = 3.88LAFGNASGTSWYY455 pKa = 9.29KK456 pKa = 9.51ACRR459 pKa = 11.84IIYY462 pKa = 7.73LTYY465 pKa = 10.01EE466 pKa = 3.78SDD468 pKa = 5.45EE469 pKa = 4.18IGEE472 pKa = 4.26WLSKK476 pKa = 10.28LVSVRR481 pKa = 11.84EE482 pKa = 3.84QDD484 pKa = 3.4VRR486 pKa = 11.84EE487 pKa = 4.12VDD489 pKa = 2.78WKK491 pKa = 10.98RR492 pKa = 11.84RR493 pKa = 11.84VEE495 pKa = 4.24SAVSSLLLNLLAGAIGGIEE514 pKa = 4.18VEE516 pKa = 4.28KK517 pKa = 10.68HH518 pKa = 4.83SCPVRR523 pKa = 11.84ASDD526 pKa = 4.95CDD528 pKa = 3.57ALSAVILMMGIRR540 pKa = 11.84QVEE543 pKa = 4.27RR544 pKa = 11.84DD545 pKa = 3.5TGGLTTWAAGRR556 pKa = 11.84IAEE559 pKa = 4.51TMTMMEE565 pKa = 4.61SKK567 pKa = 10.62VVTAIEE573 pKa = 4.16ASLIMSSGDD582 pKa = 3.25KK583 pKa = 10.82AHH585 pKa = 6.71EE586 pKa = 3.76LWANIWQRR594 pKa = 11.84GAIYY598 pKa = 10.14HH599 pKa = 5.63MPGMMWMARR608 pKa = 11.84HH609 pKa = 5.89KK610 pKa = 10.22FDD612 pKa = 4.03HH613 pKa = 6.43KK614 pKa = 10.6RR615 pKa = 11.84RR616 pKa = 11.84SDD618 pKa = 3.83GYY620 pKa = 9.51IKK622 pKa = 10.57AYY624 pKa = 9.9LARR627 pKa = 11.84LCSPKK632 pKa = 10.66SKK634 pKa = 10.42GGSGRR639 pKa = 11.84VSMTGSTFHH648 pKa = 7.53PEE650 pKa = 3.75QRR652 pKa = 11.84WSGKK656 pKa = 9.52HH657 pKa = 4.41KK658 pKa = 10.59QQGEE662 pKa = 3.85WRR664 pKa = 11.84RR665 pKa = 11.84ASGSTSLVEE674 pKa = 3.73LRR676 pKa = 11.84GIEE679 pKa = 4.09DD680 pKa = 3.82VPTISCGEE688 pKa = 4.18AIKK691 pKa = 9.46STKK694 pKa = 10.25AVALGLIGSCITQGLPAKK712 pKa = 9.5KK713 pKa = 9.12VCVVLGRR720 pKa = 11.84LLAKK724 pKa = 10.17RR725 pKa = 11.84RR726 pKa = 11.84QVLIGKK732 pKa = 7.59VEE734 pKa = 4.01KK735 pKa = 11.06VEE737 pKa = 4.48EE738 pKa = 4.51PEE740 pKa = 3.96QQLL743 pKa = 3.46
Molecular weight: 81.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3435 |
743 |
962 |
858.8 |
95.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.21 ± 0.461 | 1.456 ± 0.209 |
6.143 ± 0.511 | 6.376 ± 0.171 |
2.591 ± 0.543 | 7.54 ± 0.685 |
2.416 ± 0.298 | 4.6 ± 0.214 |
5.007 ± 0.327 | 9.054 ± 0.219 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.61 ± 0.314 | 2.969 ± 0.443 |
3.785 ± 0.406 | 2.882 ± 0.108 |
6.9 ± 0.254 | 8.21 ± 0.539 |
5.56 ± 0.288 | 7.831 ± 0.411 |
1.805 ± 0.217 | 3.057 ± 0.261 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
