
Saitoella complicata (strain BCRC 22490 / CBS 7301 / JCM 7358 / NBRC 10748 / NRRL Y-17804)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; Taphrinomycotina; Taphrinomycotina incertae sedis; Saitoella; Saitoella complicata
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7118 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E9N7Z9|A0A0E9N7Z9_SAICN Uncharacterized protein OS=Saitoella complicata (strain BCRC 22490 / CBS 7301 / JCM 7358 / NBRC 10748 / NRRL Y-17804) OX=698492 GN=G7K_0095-t2 PE=4 SV=1
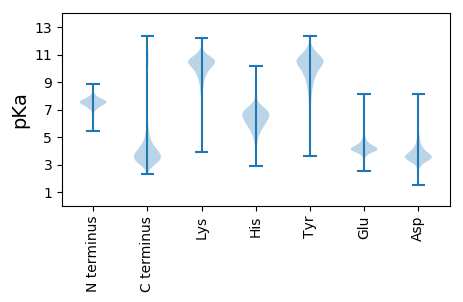
MM1 pKa = 7.37VNGSFYY7 pKa = 10.94GPQPQRR13 pKa = 11.84TAGVGSYY20 pKa = 10.92SPVAVVNMSEE30 pKa = 4.44SEE32 pKa = 4.06PSQALYY38 pKa = 10.56YY39 pKa = 10.59CHH41 pKa = 7.09EE42 pKa = 5.02CQDD45 pKa = 3.06EE46 pKa = 4.28WNAQEE51 pKa = 4.51TDD53 pKa = 2.87VGMPCPRR60 pKa = 11.84CGSDD64 pKa = 3.19FTEE67 pKa = 5.45VIDD70 pKa = 4.55QGADD74 pKa = 3.38PRR76 pKa = 11.84DD77 pKa = 3.76LLHH80 pKa = 8.02DD81 pKa = 5.16DD82 pKa = 4.51GDD84 pKa = 4.12DD85 pKa = 4.26FEE87 pKa = 8.01DD88 pKa = 5.07DD89 pKa = 5.33DD90 pKa = 6.06EE91 pKa = 6.7YY92 pKa = 11.78DD93 pKa = 4.07DD94 pKa = 6.46DD95 pKa = 5.35EE96 pKa = 6.5DD97 pKa = 4.92VDD99 pKa = 4.22GLGFNVAGPGGMPPFMFGNIAGNNTDD125 pKa = 3.49HH126 pKa = 6.72GPNRR130 pKa = 11.84EE131 pKa = 4.02MMGMLQGLLQQFIGGRR147 pKa = 11.84QPPPRR152 pKa = 11.84ADD154 pKa = 3.49SNEE157 pKa = 4.0NGHH160 pKa = 5.26TTSVDD165 pKa = 3.24MPAQPQQAAGANVQFTFGSMAAGLGVAGTGRR196 pKa = 11.84GDD198 pKa = 3.39RR199 pKa = 11.84QEE201 pKa = 4.3NGSGHH206 pKa = 6.1VPVTDD211 pKa = 4.69LSTFLQQAFGALAGGPADD229 pKa = 3.73GGFAFGGGNGQAGGGLGRR247 pKa = 11.84NLGDD251 pKa = 3.16YY252 pKa = 10.2VFSNRR257 pKa = 11.84GFDD260 pKa = 3.53EE261 pKa = 4.43VVSRR265 pKa = 11.84LMEE268 pKa = 3.71QHH270 pKa = 6.16QGSDD274 pKa = 3.67APPPTPEE281 pKa = 3.89YY282 pKa = 10.56VINAIPAFLVDD293 pKa = 3.62EE294 pKa = 5.22NIAKK298 pKa = 10.16SEE300 pKa = 3.95WDD302 pKa = 3.62CAVCKK307 pKa = 10.46DD308 pKa = 3.52DD309 pKa = 3.6WQIGEE314 pKa = 4.11TALRR318 pKa = 11.84LPCSHH323 pKa = 7.02CFHH326 pKa = 7.15DD327 pKa = 4.32QCIKK331 pKa = 10.47PWLKK335 pKa = 10.93VNSTCPVCRR344 pKa = 11.84SSVLGSGTAQRR355 pKa = 11.84SAEE358 pKa = 4.01QAGGSSVEE366 pKa = 4.28GVTVATQASSSTSHH380 pKa = 5.79QMPGSYY386 pKa = 9.14PASASAPDD394 pKa = 3.88ILEE397 pKa = 4.2QDD399 pKa = 4.28DD400 pKa = 4.34LDD402 pKa = 3.85
MM1 pKa = 7.37VNGSFYY7 pKa = 10.94GPQPQRR13 pKa = 11.84TAGVGSYY20 pKa = 10.92SPVAVVNMSEE30 pKa = 4.44SEE32 pKa = 4.06PSQALYY38 pKa = 10.56YY39 pKa = 10.59CHH41 pKa = 7.09EE42 pKa = 5.02CQDD45 pKa = 3.06EE46 pKa = 4.28WNAQEE51 pKa = 4.51TDD53 pKa = 2.87VGMPCPRR60 pKa = 11.84CGSDD64 pKa = 3.19FTEE67 pKa = 5.45VIDD70 pKa = 4.55QGADD74 pKa = 3.38PRR76 pKa = 11.84DD77 pKa = 3.76LLHH80 pKa = 8.02DD81 pKa = 5.16DD82 pKa = 4.51GDD84 pKa = 4.12DD85 pKa = 4.26FEE87 pKa = 8.01DD88 pKa = 5.07DD89 pKa = 5.33DD90 pKa = 6.06EE91 pKa = 6.7YY92 pKa = 11.78DD93 pKa = 4.07DD94 pKa = 6.46DD95 pKa = 5.35EE96 pKa = 6.5DD97 pKa = 4.92VDD99 pKa = 4.22GLGFNVAGPGGMPPFMFGNIAGNNTDD125 pKa = 3.49HH126 pKa = 6.72GPNRR130 pKa = 11.84EE131 pKa = 4.02MMGMLQGLLQQFIGGRR147 pKa = 11.84QPPPRR152 pKa = 11.84ADD154 pKa = 3.49SNEE157 pKa = 4.0NGHH160 pKa = 5.26TTSVDD165 pKa = 3.24MPAQPQQAAGANVQFTFGSMAAGLGVAGTGRR196 pKa = 11.84GDD198 pKa = 3.39RR199 pKa = 11.84QEE201 pKa = 4.3NGSGHH206 pKa = 6.1VPVTDD211 pKa = 4.69LSTFLQQAFGALAGGPADD229 pKa = 3.73GGFAFGGGNGQAGGGLGRR247 pKa = 11.84NLGDD251 pKa = 3.16YY252 pKa = 10.2VFSNRR257 pKa = 11.84GFDD260 pKa = 3.53EE261 pKa = 4.43VVSRR265 pKa = 11.84LMEE268 pKa = 3.71QHH270 pKa = 6.16QGSDD274 pKa = 3.67APPPTPEE281 pKa = 3.89YY282 pKa = 10.56VINAIPAFLVDD293 pKa = 3.62EE294 pKa = 5.22NIAKK298 pKa = 10.16SEE300 pKa = 3.95WDD302 pKa = 3.62CAVCKK307 pKa = 10.46DD308 pKa = 3.52DD309 pKa = 3.6WQIGEE314 pKa = 4.11TALRR318 pKa = 11.84LPCSHH323 pKa = 7.02CFHH326 pKa = 7.15DD327 pKa = 4.32QCIKK331 pKa = 10.47PWLKK335 pKa = 10.93VNSTCPVCRR344 pKa = 11.84SSVLGSGTAQRR355 pKa = 11.84SAEE358 pKa = 4.01QAGGSSVEE366 pKa = 4.28GVTVATQASSSTSHH380 pKa = 5.79QMPGSYY386 pKa = 9.14PASASAPDD394 pKa = 3.88ILEE397 pKa = 4.2QDD399 pKa = 4.28DD400 pKa = 4.34LDD402 pKa = 3.85
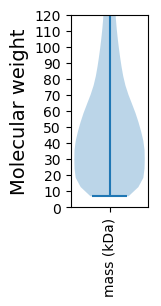
Molecular weight: 42.3 kDa
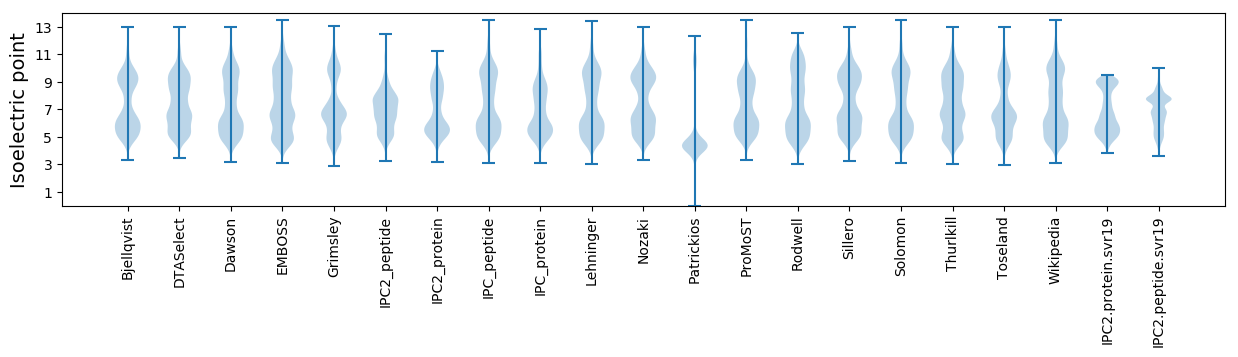
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E9NKP6|A0A0E9NKP6_SAICN Sm domain-containing protein OS=Saitoella complicata (strain BCRC 22490 / CBS 7301 / JCM 7358 / NBRC 10748 / NRRL Y-17804) OX=698492 GN=G7K_4115-t1 PE=3 SV=1
MM1 pKa = 7.87RR2 pKa = 11.84GNKK5 pKa = 8.53VRR7 pKa = 11.84LLPALPLSRR16 pKa = 11.84LRR18 pKa = 11.84ILNLKK23 pKa = 7.7TLHH26 pKa = 6.66LSTSFHH32 pKa = 6.69PLSGPSNLNRR42 pKa = 11.84TTPAAVPRR50 pKa = 11.84PWRR53 pKa = 11.84GRR55 pKa = 11.84LHH57 pKa = 6.38SVPLRR62 pKa = 11.84LRR64 pKa = 11.84QPLTHH69 pKa = 7.04RR70 pKa = 4.3
MM1 pKa = 7.87RR2 pKa = 11.84GNKK5 pKa = 8.53VRR7 pKa = 11.84LLPALPLSRR16 pKa = 11.84LRR18 pKa = 11.84ILNLKK23 pKa = 7.7TLHH26 pKa = 6.66LSTSFHH32 pKa = 6.69PLSGPSNLNRR42 pKa = 11.84TTPAAVPRR50 pKa = 11.84PWRR53 pKa = 11.84GRR55 pKa = 11.84LHH57 pKa = 6.38SVPLRR62 pKa = 11.84LRR64 pKa = 11.84QPLTHH69 pKa = 7.04RR70 pKa = 4.3
Molecular weight: 7.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3658384 |
66 |
5171 |
514.0 |
56.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.74 ± 0.028 | 1.267 ± 0.009 |
5.311 ± 0.018 | 6.729 ± 0.029 |
3.529 ± 0.017 | 6.99 ± 0.026 |
2.414 ± 0.013 | 4.62 ± 0.018 |
5.04 ± 0.022 | 8.85 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.303 ± 0.009 | 3.313 ± 0.015 |
6.015 ± 0.033 | 3.729 ± 0.019 |
6.336 ± 0.029 | 7.896 ± 0.033 |
6.228 ± 0.024 | 6.681 ± 0.019 |
1.338 ± 0.011 | 2.67 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
