
Candidatus Entotheonella factor
Taxonomy: cellular organisms; Bacteria; Nitrospinae/Tectomicrobia group; Candidatus Tectomicrobia; Candidatus Entotheonella
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
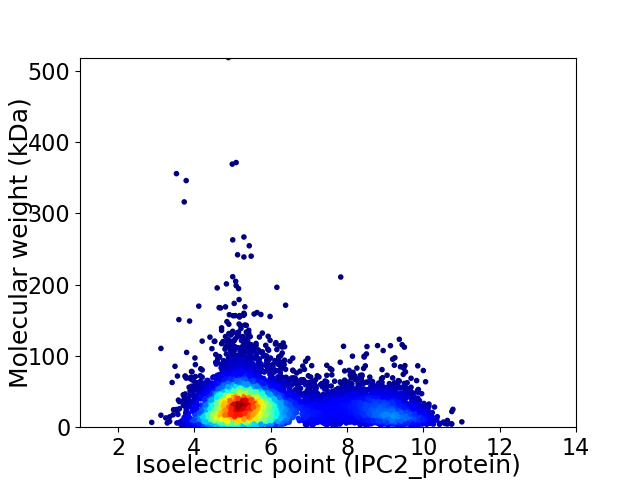
Virtual 2D-PAGE plot for 8429 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W4LAU2|W4LAU2_9BACT Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex OS=Candidatus Entotheonella factor OX=1429438 GN=ETSY1_32265 PE=3 SV=1
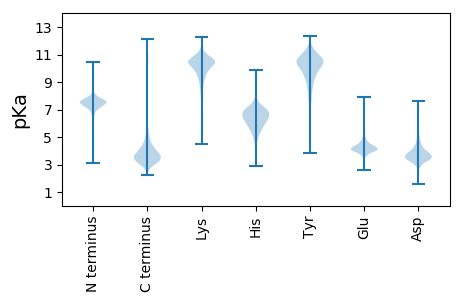
MM1 pKa = 7.52LLSLNTIFTWCLLVAWLVKK20 pKa = 10.41PFGFTADD27 pKa = 3.58AQALLPSDD35 pKa = 4.12VQTVDD40 pKa = 2.93ATGDD44 pKa = 3.5TMVNLTLPHH53 pKa = 6.83GFTLSGAVRR62 pKa = 11.84TEE64 pKa = 3.78RR65 pKa = 11.84GDD67 pKa = 3.65PVALGTLTARR77 pKa = 11.84STGGQFTSSLQGGDD91 pKa = 3.46SAYY94 pKa = 10.62HH95 pKa = 5.98IALPGGTYY103 pKa = 10.67DD104 pKa = 4.65LNLSIPFLEE113 pKa = 4.43SDD115 pKa = 3.49EE116 pKa = 4.27TGIPIFVYY124 pKa = 9.31MASDD128 pKa = 3.77VVAGLGIAADD138 pKa = 3.95TTQDD142 pKa = 3.79LVVPDD147 pKa = 4.86LPDD150 pKa = 3.43LVTVTGFVDD159 pKa = 3.74PQDD162 pKa = 3.92ADD164 pKa = 3.92TVPTEE169 pKa = 4.07GALQLVSTDD178 pKa = 3.04GGTFTLALFEE188 pKa = 5.09DD189 pKa = 6.15FYY191 pKa = 10.01TTRR194 pKa = 11.84LPLNTYY200 pKa = 9.35NVSAALTFTEE210 pKa = 5.42AIGTNGPRR218 pKa = 11.84TTAKK222 pKa = 10.66VIVNVDD228 pKa = 3.01AVTVNDD234 pKa = 4.09EE235 pKa = 3.89PTIYY239 pKa = 10.58DD240 pKa = 3.14ITLPATANLSGTVLDD255 pKa = 4.33SAAAPLAPARR265 pKa = 11.84VVATAVDD272 pKa = 3.38ADD274 pKa = 3.86AAVPSDD280 pKa = 3.19IDD282 pKa = 3.77FSCRR286 pKa = 11.84PEE288 pKa = 4.02VTFSLVPMPVSGTAFLYY305 pKa = 10.49RR306 pKa = 11.84EE307 pKa = 4.47PGEE310 pKa = 4.27SSTVGAYY317 pKa = 10.04HH318 pKa = 6.42MPLPIGTYY326 pKa = 9.69HH327 pKa = 7.29LNVTLGLEE335 pKa = 4.4LQPEE339 pKa = 4.43MVPTLVDD346 pKa = 3.64PEE348 pKa = 4.52VSSSSIVHH356 pKa = 6.24IAMPDD361 pKa = 3.27VALTTDD367 pKa = 3.81TAQNLAVPALPPVVMVSGLVTDD389 pKa = 5.15ALGQPVANAAVVAMTEE405 pKa = 3.94ALAGVANAVRR415 pKa = 11.84FLAEE419 pKa = 3.9VQTQEE424 pKa = 4.23DD425 pKa = 3.98GSYY428 pKa = 10.1EE429 pKa = 4.13LPVLSGTSYY438 pKa = 11.05TIMACPPRR446 pKa = 11.84LQQ448 pKa = 3.45
MM1 pKa = 7.52LLSLNTIFTWCLLVAWLVKK20 pKa = 10.41PFGFTADD27 pKa = 3.58AQALLPSDD35 pKa = 4.12VQTVDD40 pKa = 2.93ATGDD44 pKa = 3.5TMVNLTLPHH53 pKa = 6.83GFTLSGAVRR62 pKa = 11.84TEE64 pKa = 3.78RR65 pKa = 11.84GDD67 pKa = 3.65PVALGTLTARR77 pKa = 11.84STGGQFTSSLQGGDD91 pKa = 3.46SAYY94 pKa = 10.62HH95 pKa = 5.98IALPGGTYY103 pKa = 10.67DD104 pKa = 4.65LNLSIPFLEE113 pKa = 4.43SDD115 pKa = 3.49EE116 pKa = 4.27TGIPIFVYY124 pKa = 9.31MASDD128 pKa = 3.77VVAGLGIAADD138 pKa = 3.95TTQDD142 pKa = 3.79LVVPDD147 pKa = 4.86LPDD150 pKa = 3.43LVTVTGFVDD159 pKa = 3.74PQDD162 pKa = 3.92ADD164 pKa = 3.92TVPTEE169 pKa = 4.07GALQLVSTDD178 pKa = 3.04GGTFTLALFEE188 pKa = 5.09DD189 pKa = 6.15FYY191 pKa = 10.01TTRR194 pKa = 11.84LPLNTYY200 pKa = 9.35NVSAALTFTEE210 pKa = 5.42AIGTNGPRR218 pKa = 11.84TTAKK222 pKa = 10.66VIVNVDD228 pKa = 3.01AVTVNDD234 pKa = 4.09EE235 pKa = 3.89PTIYY239 pKa = 10.58DD240 pKa = 3.14ITLPATANLSGTVLDD255 pKa = 4.33SAAAPLAPARR265 pKa = 11.84VVATAVDD272 pKa = 3.38ADD274 pKa = 3.86AAVPSDD280 pKa = 3.19IDD282 pKa = 3.77FSCRR286 pKa = 11.84PEE288 pKa = 4.02VTFSLVPMPVSGTAFLYY305 pKa = 10.49RR306 pKa = 11.84EE307 pKa = 4.47PGEE310 pKa = 4.27SSTVGAYY317 pKa = 10.04HH318 pKa = 6.42MPLPIGTYY326 pKa = 9.69HH327 pKa = 7.29LNVTLGLEE335 pKa = 4.4LQPEE339 pKa = 4.43MVPTLVDD346 pKa = 3.64PEE348 pKa = 4.52VSSSSIVHH356 pKa = 6.24IAMPDD361 pKa = 3.27VALTTDD367 pKa = 3.81TAQNLAVPALPPVVMVSGLVTDD389 pKa = 5.15ALGQPVANAAVVAMTEE405 pKa = 3.94ALAGVANAVRR415 pKa = 11.84FLAEE419 pKa = 3.9VQTQEE424 pKa = 4.23DD425 pKa = 3.98GSYY428 pKa = 10.1EE429 pKa = 4.13LPVLSGTSYY438 pKa = 11.05TIMACPPRR446 pKa = 11.84LQQ448 pKa = 3.45
Molecular weight: 46.62 kDa
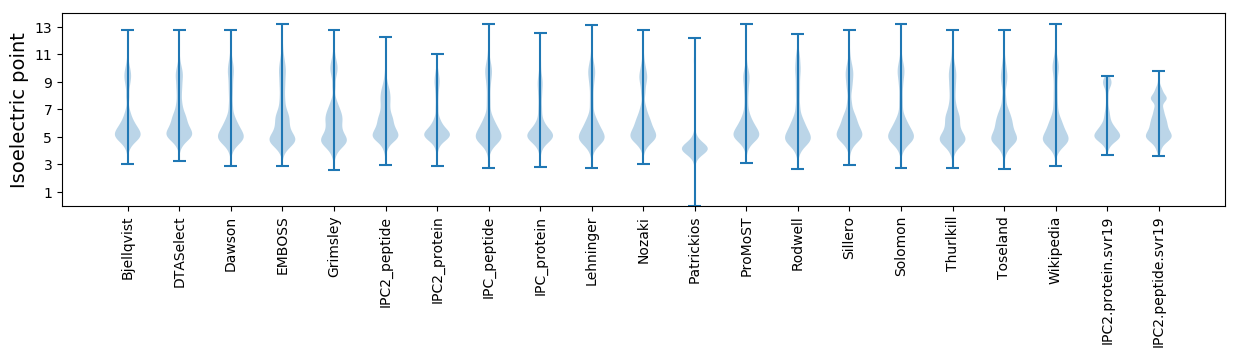
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W4LII1|W4LII1_9BACT Uncharacterized protein OS=Candidatus Entotheonella factor OX=1429438 GN=ETSY1_21555 PE=4 SV=1
MM1 pKa = 7.89PKK3 pKa = 9.71MKK5 pKa = 9.95TNRR8 pKa = 11.84SAAKK12 pKa = 10.05RR13 pKa = 11.84FRR15 pKa = 11.84LTARR19 pKa = 11.84GKK21 pKa = 9.33VRR23 pKa = 11.84RR24 pKa = 11.84NQAFTRR30 pKa = 11.84HH31 pKa = 5.85ILTKK35 pKa = 9.37KK36 pKa = 6.62TRR38 pKa = 11.84KK39 pKa = 9.46RR40 pKa = 11.84KK41 pKa = 8.59RR42 pKa = 11.84QLRR45 pKa = 11.84STTLVAAADD54 pKa = 3.59APRR57 pKa = 11.84IKK59 pKa = 10.35RR60 pKa = 11.84IISSS64 pKa = 3.44
MM1 pKa = 7.89PKK3 pKa = 9.71MKK5 pKa = 9.95TNRR8 pKa = 11.84SAAKK12 pKa = 10.05RR13 pKa = 11.84FRR15 pKa = 11.84LTARR19 pKa = 11.84GKK21 pKa = 9.33VRR23 pKa = 11.84RR24 pKa = 11.84NQAFTRR30 pKa = 11.84HH31 pKa = 5.85ILTKK35 pKa = 9.37KK36 pKa = 6.62TRR38 pKa = 11.84KK39 pKa = 9.46RR40 pKa = 11.84KK41 pKa = 8.59RR42 pKa = 11.84QLRR45 pKa = 11.84STTLVAAADD54 pKa = 3.59APRR57 pKa = 11.84IKK59 pKa = 10.35RR60 pKa = 11.84IISSS64 pKa = 3.44
Molecular weight: 7.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2493613 |
29 |
4723 |
295.8 |
32.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.08 ± 0.028 | 1.07 ± 0.01 |
5.707 ± 0.021 | 6.085 ± 0.028 |
3.62 ± 0.016 | 7.591 ± 0.03 |
2.74 ± 0.014 | 5.275 ± 0.021 |
2.854 ± 0.021 | 10.487 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.573 ± 0.015 | 2.769 ± 0.018 |
5.327 ± 0.02 | 4.668 ± 0.026 |
6.685 ± 0.025 | 5.367 ± 0.017 |
5.535 ± 0.024 | 7.183 ± 0.023 |
1.505 ± 0.013 | 2.878 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
