
Toros virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Toros phlebovirus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
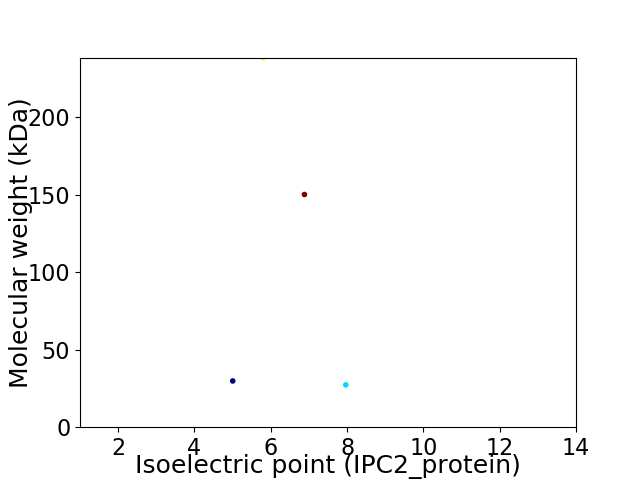
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A161D6V4|A0A161D6V4_9VIRU Viral sRNA Nonstructural protein OS=Toros virus OX=1764085 PE=4 SV=1
MM1 pKa = 7.39NSQYY5 pKa = 11.17LFDD8 pKa = 4.25YY9 pKa = 10.4PVVKK13 pKa = 10.4LDD15 pKa = 3.79SRR17 pKa = 11.84VHH19 pKa = 6.16RR20 pKa = 11.84ALSTVNYY27 pKa = 7.98MAFNKK32 pKa = 9.54FHH34 pKa = 6.64YY35 pKa = 9.91CDD37 pKa = 3.14ISTYY41 pKa = 6.86EE42 pKa = 3.75HH43 pKa = 7.22CEE45 pKa = 3.93FPLEE49 pKa = 4.28KK50 pKa = 10.24YY51 pKa = 10.44SRR53 pKa = 11.84GFGRR57 pKa = 11.84RR58 pKa = 11.84GSLSDD63 pKa = 3.84FYY65 pKa = 11.23LYY67 pKa = 10.82KK68 pKa = 10.4EE69 pKa = 5.11LPASWGPACTISSVKK84 pKa = 10.17PIVYY88 pKa = 9.43PFRR91 pKa = 11.84GLANDD96 pKa = 4.58LSQFDD101 pKa = 3.97MQSFSVPGLQNIRR114 pKa = 11.84KK115 pKa = 8.76ALSWPFGFPDD125 pKa = 3.88LEE127 pKa = 4.02IFEE130 pKa = 4.71ICSEE134 pKa = 4.61GYY136 pKa = 10.11KK137 pKa = 10.35RR138 pKa = 11.84GLDD141 pKa = 3.41TRR143 pKa = 11.84NQLMSYY149 pKa = 8.87ILRR152 pKa = 11.84MADD155 pKa = 3.1SKK157 pKa = 11.78YY158 pKa = 10.57LDD160 pKa = 3.48EE161 pKa = 6.44CIVQAHH167 pKa = 6.19KK168 pKa = 10.71KK169 pKa = 9.63ILSEE173 pKa = 4.13CRR175 pKa = 11.84ALGLQDD181 pKa = 3.43EE182 pKa = 4.98HH183 pKa = 7.89FNGYY187 pKa = 10.23DD188 pKa = 3.38LFRR191 pKa = 11.84EE192 pKa = 4.11ISTLVCTRR200 pKa = 11.84LINAEE205 pKa = 4.45PYY207 pKa = 10.61DD208 pKa = 4.03STSSGSGLEE217 pKa = 3.77IKK219 pKa = 10.58CIIRR223 pKa = 11.84SYY225 pKa = 10.42KK226 pKa = 10.04ISDD229 pKa = 3.53PSSCVGVYY237 pKa = 10.59GSGFWEE243 pKa = 4.15PVNEE247 pKa = 4.25PEE249 pKa = 4.15EE250 pKa = 4.2DD251 pKa = 3.68TYY253 pKa = 11.8EE254 pKa = 4.81DD255 pKa = 4.41PDD257 pKa = 4.25SEE259 pKa = 4.47FF260 pKa = 3.9
MM1 pKa = 7.39NSQYY5 pKa = 11.17LFDD8 pKa = 4.25YY9 pKa = 10.4PVVKK13 pKa = 10.4LDD15 pKa = 3.79SRR17 pKa = 11.84VHH19 pKa = 6.16RR20 pKa = 11.84ALSTVNYY27 pKa = 7.98MAFNKK32 pKa = 9.54FHH34 pKa = 6.64YY35 pKa = 9.91CDD37 pKa = 3.14ISTYY41 pKa = 6.86EE42 pKa = 3.75HH43 pKa = 7.22CEE45 pKa = 3.93FPLEE49 pKa = 4.28KK50 pKa = 10.24YY51 pKa = 10.44SRR53 pKa = 11.84GFGRR57 pKa = 11.84RR58 pKa = 11.84GSLSDD63 pKa = 3.84FYY65 pKa = 11.23LYY67 pKa = 10.82KK68 pKa = 10.4EE69 pKa = 5.11LPASWGPACTISSVKK84 pKa = 10.17PIVYY88 pKa = 9.43PFRR91 pKa = 11.84GLANDD96 pKa = 4.58LSQFDD101 pKa = 3.97MQSFSVPGLQNIRR114 pKa = 11.84KK115 pKa = 8.76ALSWPFGFPDD125 pKa = 3.88LEE127 pKa = 4.02IFEE130 pKa = 4.71ICSEE134 pKa = 4.61GYY136 pKa = 10.11KK137 pKa = 10.35RR138 pKa = 11.84GLDD141 pKa = 3.41TRR143 pKa = 11.84NQLMSYY149 pKa = 8.87ILRR152 pKa = 11.84MADD155 pKa = 3.1SKK157 pKa = 11.78YY158 pKa = 10.57LDD160 pKa = 3.48EE161 pKa = 6.44CIVQAHH167 pKa = 6.19KK168 pKa = 10.71KK169 pKa = 9.63ILSEE173 pKa = 4.13CRR175 pKa = 11.84ALGLQDD181 pKa = 3.43EE182 pKa = 4.98HH183 pKa = 7.89FNGYY187 pKa = 10.23DD188 pKa = 3.38LFRR191 pKa = 11.84EE192 pKa = 4.11ISTLVCTRR200 pKa = 11.84LINAEE205 pKa = 4.45PYY207 pKa = 10.61DD208 pKa = 4.03STSSGSGLEE217 pKa = 3.77IKK219 pKa = 10.58CIIRR223 pKa = 11.84SYY225 pKa = 10.42KK226 pKa = 10.04ISDD229 pKa = 3.53PSSCVGVYY237 pKa = 10.59GSGFWEE243 pKa = 4.15PVNEE247 pKa = 4.25PEE249 pKa = 4.15EE250 pKa = 4.2DD251 pKa = 3.68TYY253 pKa = 11.8EE254 pKa = 4.81DD255 pKa = 4.41PDD257 pKa = 4.25SEE259 pKa = 4.47FF260 pKa = 3.9
Molecular weight: 29.84 kDa
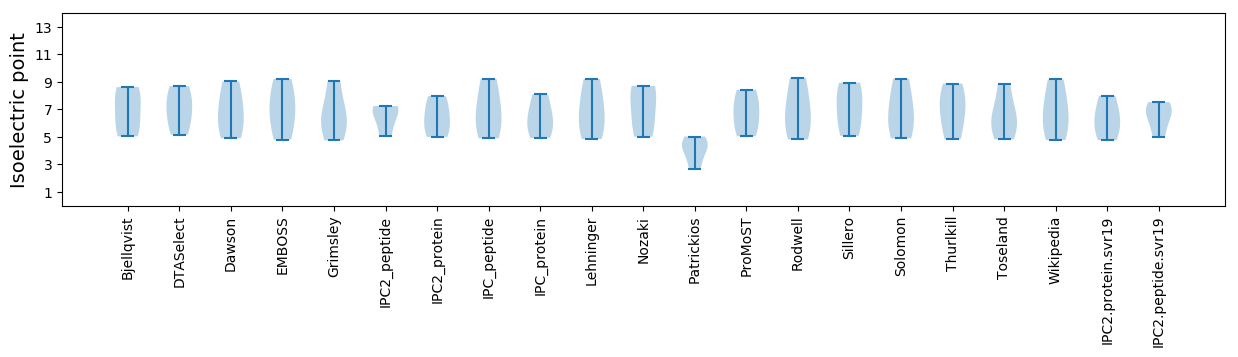
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161D6V4|A0A161D6V4_9VIRU Viral sRNA Nonstructural protein OS=Toros virus OX=1764085 PE=4 SV=1
MM1 pKa = 7.89EE2 pKa = 6.57DD3 pKa = 3.44YY4 pKa = 11.12QKK6 pKa = 10.78IAVEE10 pKa = 4.38FGEE13 pKa = 4.06QAIDD17 pKa = 3.46EE18 pKa = 4.69TVVQEE23 pKa = 3.8WLQNFAYY30 pKa = 10.29QGFDD34 pKa = 2.91ARR36 pKa = 11.84AVIQNLIKK44 pKa = 10.84LGGSSWEE51 pKa = 3.83EE52 pKa = 3.78DD53 pKa = 3.27AKK55 pKa = 11.55KK56 pKa = 10.12MIILALTRR64 pKa = 11.84GNKK67 pKa = 7.62PKK69 pKa = 10.62KK70 pKa = 7.39MQEE73 pKa = 3.72RR74 pKa = 11.84MSPEE78 pKa = 3.43GAKK81 pKa = 9.58EE82 pKa = 4.01VKK84 pKa = 10.63ALVAKK89 pKa = 10.31YY90 pKa = 10.21KK91 pKa = 10.6LVEE94 pKa = 4.38GKK96 pKa = 9.54PGRR99 pKa = 11.84NGLTLTRR106 pKa = 11.84VAAALAGWTVQAIEE120 pKa = 4.28VVEE123 pKa = 4.23NDD125 pKa = 3.58LPVPGSAMDD134 pKa = 4.86RR135 pKa = 11.84ISGQTYY141 pKa = 7.91PRR143 pKa = 11.84QMMHH147 pKa = 6.99PSFAGLIDD155 pKa = 4.38PSLDD159 pKa = 3.45PEE161 pKa = 4.54DD162 pKa = 4.26FNAIVDD168 pKa = 3.57AHH170 pKa = 6.92KK171 pKa = 10.86LFLYY175 pKa = 8.85MFSKK179 pKa = 9.59TINVGLRR186 pKa = 11.84GASKK190 pKa = 10.7RR191 pKa = 11.84DD192 pKa = 3.27IEE194 pKa = 4.43EE195 pKa = 4.39SFSQPMSAAINSSFIDD211 pKa = 3.35NAQRR215 pKa = 11.84RR216 pKa = 11.84SFLTKK221 pKa = 9.89FSIVTSSSRR230 pKa = 11.84ASSVVKK236 pKa = 10.49KK237 pKa = 10.21IAEE240 pKa = 4.22VYY242 pKa = 10.2RR243 pKa = 11.84KK244 pKa = 10.34LEE246 pKa = 3.85
MM1 pKa = 7.89EE2 pKa = 6.57DD3 pKa = 3.44YY4 pKa = 11.12QKK6 pKa = 10.78IAVEE10 pKa = 4.38FGEE13 pKa = 4.06QAIDD17 pKa = 3.46EE18 pKa = 4.69TVVQEE23 pKa = 3.8WLQNFAYY30 pKa = 10.29QGFDD34 pKa = 2.91ARR36 pKa = 11.84AVIQNLIKK44 pKa = 10.84LGGSSWEE51 pKa = 3.83EE52 pKa = 3.78DD53 pKa = 3.27AKK55 pKa = 11.55KK56 pKa = 10.12MIILALTRR64 pKa = 11.84GNKK67 pKa = 7.62PKK69 pKa = 10.62KK70 pKa = 7.39MQEE73 pKa = 3.72RR74 pKa = 11.84MSPEE78 pKa = 3.43GAKK81 pKa = 9.58EE82 pKa = 4.01VKK84 pKa = 10.63ALVAKK89 pKa = 10.31YY90 pKa = 10.21KK91 pKa = 10.6LVEE94 pKa = 4.38GKK96 pKa = 9.54PGRR99 pKa = 11.84NGLTLTRR106 pKa = 11.84VAAALAGWTVQAIEE120 pKa = 4.28VVEE123 pKa = 4.23NDD125 pKa = 3.58LPVPGSAMDD134 pKa = 4.86RR135 pKa = 11.84ISGQTYY141 pKa = 7.91PRR143 pKa = 11.84QMMHH147 pKa = 6.99PSFAGLIDD155 pKa = 4.38PSLDD159 pKa = 3.45PEE161 pKa = 4.54DD162 pKa = 4.26FNAIVDD168 pKa = 3.57AHH170 pKa = 6.92KK171 pKa = 10.86LFLYY175 pKa = 8.85MFSKK179 pKa = 9.59TINVGLRR186 pKa = 11.84GASKK190 pKa = 10.7RR191 pKa = 11.84DD192 pKa = 3.27IEE194 pKa = 4.43EE195 pKa = 4.39SFSQPMSAAINSSFIDD211 pKa = 3.35NAQRR215 pKa = 11.84RR216 pKa = 11.84SFLTKK221 pKa = 9.89FSIVTSSSRR230 pKa = 11.84ASSVVKK236 pKa = 10.49KK237 pKa = 10.21IAEE240 pKa = 4.22VYY242 pKa = 10.2RR243 pKa = 11.84KK244 pKa = 10.34LEE246 pKa = 3.85
Molecular weight: 27.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3955 |
246 |
2090 |
988.8 |
111.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.917 ± 0.664 | 2.528 ± 0.85 |
5.588 ± 0.325 | 7.434 ± 0.373 |
4.399 ± 0.491 | 6.119 ± 0.032 |
2.048 ± 0.177 | 6.068 ± 0.276 |
6.574 ± 0.369 | 8.47 ± 0.473 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.363 ± 0.271 | 4.02 ± 0.19 |
3.692 ± 0.382 | 2.958 ± 0.406 |
5.74 ± 0.513 | 8.723 ± 0.52 |
5.386 ± 0.574 | 6.802 ± 0.334 |
1.214 ± 0.058 | 2.958 ± 0.559 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
