
Bacillus lehensis G1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Alkalihalobacillus; Alkalihalobacillus lehensis
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
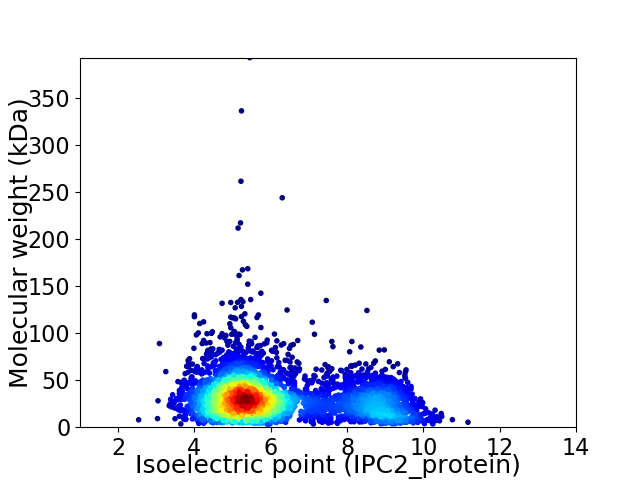
Virtual 2D-PAGE plot for 4016 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
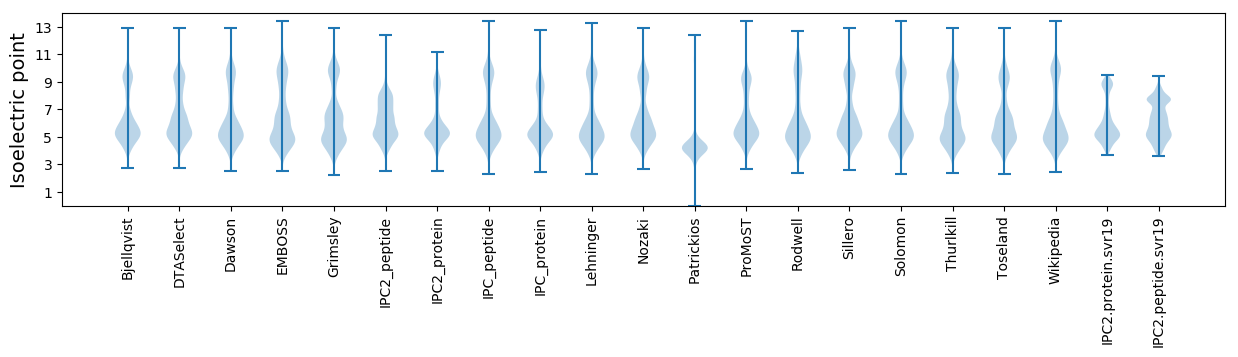
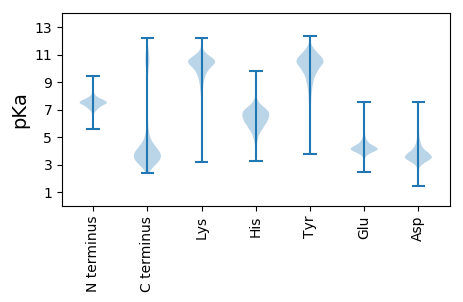
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A060M758|A0A060M758_9BACI Glycosyltransferase OS=Bacillus lehensis G1 OX=1246626 GN=BleG1_3372 PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.24KK3 pKa = 9.75VFGVGFVSALAITTLAACGSGSDD26 pKa = 4.56SSTGTNGDD34 pKa = 3.5VVEE37 pKa = 5.28LSFSAWGNPAEE48 pKa = 4.05LDD50 pKa = 3.77VYY52 pKa = 11.09NRR54 pKa = 11.84AVDD57 pKa = 4.08AFNEE61 pKa = 4.29MQDD64 pKa = 3.84DD65 pKa = 4.24VQVTLTGLPNDD76 pKa = 4.44NYY78 pKa = 10.55FQTLTTRR85 pKa = 11.84LQGGQAPDD93 pKa = 3.27VFYY96 pKa = 11.45VGAEE100 pKa = 4.13NISTLMSNGTIEE112 pKa = 4.23PLSDD116 pKa = 3.5FLEE119 pKa = 5.08SGDD122 pKa = 4.02SYY124 pKa = 10.28VTEE127 pKa = 4.98DD128 pKa = 4.84EE129 pKa = 4.2FTEE132 pKa = 4.97DD133 pKa = 1.97IWGASRR139 pKa = 11.84NDD141 pKa = 4.18GIVYY145 pKa = 8.9GLPVDD150 pKa = 4.31CNPILMYY157 pKa = 10.51YY158 pKa = 10.42NKK160 pKa = 10.76ALLEE164 pKa = 4.28EE165 pKa = 4.48NNIPSPQEE173 pKa = 3.56LYY175 pKa = 11.21EE176 pKa = 4.26NGEE179 pKa = 4.4WNWDD183 pKa = 3.47TFEE186 pKa = 6.91DD187 pKa = 3.55ITGQLNEE194 pKa = 4.7SGNHH198 pKa = 5.96GYY200 pKa = 7.92MQEE203 pKa = 4.17SGHH206 pKa = 5.99GAMLNWIWTNGGEE219 pKa = 4.67FATEE223 pKa = 3.73DD224 pKa = 4.45AIVGDD229 pKa = 4.06TDD231 pKa = 3.69GKK233 pKa = 8.98TMEE236 pKa = 4.24AFEE239 pKa = 4.32YY240 pKa = 8.64VDD242 pKa = 4.56RR243 pKa = 11.84MIDD246 pKa = 3.38EE247 pKa = 5.1GNFTYY252 pKa = 10.6AGTMPEE258 pKa = 4.21GQGIEE263 pKa = 4.09AMFMSNQVGFVSAGRR278 pKa = 11.84WLTPMFLDD286 pKa = 3.71TSIDD290 pKa = 3.24FDD292 pKa = 4.79YY293 pKa = 11.26IPFPSNTDD301 pKa = 3.43EE302 pKa = 4.7QIEE305 pKa = 4.28TANIATAYY313 pKa = 8.55MAVNSKK319 pKa = 10.44SDD321 pKa = 3.46HH322 pKa = 6.46VEE324 pKa = 3.46EE325 pKa = 4.04AMKK328 pKa = 10.54FISYY332 pKa = 7.59YY333 pKa = 10.19TSTDD337 pKa = 3.32GQEE340 pKa = 3.6VRR342 pKa = 11.84LDD344 pKa = 3.76GEE346 pKa = 4.75GNAVPSVVGIDD357 pKa = 3.83EE358 pKa = 4.15IVLNQDD364 pKa = 3.18RR365 pKa = 11.84PEE367 pKa = 4.52HH368 pKa = 5.43GQYY371 pKa = 11.25LLDD374 pKa = 3.88ARR376 pKa = 11.84SVGRR380 pKa = 11.84VIGLEE385 pKa = 3.98FTSPGLSEE393 pKa = 4.7DD394 pKa = 4.22LEE396 pKa = 4.95DD397 pKa = 4.29IYY399 pKa = 11.34EE400 pKa = 4.25LMMLGDD406 pKa = 4.16IDD408 pKa = 5.54AGDD411 pKa = 4.32ALEE414 pKa = 4.19QATAKK419 pKa = 10.65AKK421 pKa = 10.61AAVEE425 pKa = 4.11
MM1 pKa = 7.42KK2 pKa = 10.24KK3 pKa = 9.75VFGVGFVSALAITTLAACGSGSDD26 pKa = 4.56SSTGTNGDD34 pKa = 3.5VVEE37 pKa = 5.28LSFSAWGNPAEE48 pKa = 4.05LDD50 pKa = 3.77VYY52 pKa = 11.09NRR54 pKa = 11.84AVDD57 pKa = 4.08AFNEE61 pKa = 4.29MQDD64 pKa = 3.84DD65 pKa = 4.24VQVTLTGLPNDD76 pKa = 4.44NYY78 pKa = 10.55FQTLTTRR85 pKa = 11.84LQGGQAPDD93 pKa = 3.27VFYY96 pKa = 11.45VGAEE100 pKa = 4.13NISTLMSNGTIEE112 pKa = 4.23PLSDD116 pKa = 3.5FLEE119 pKa = 5.08SGDD122 pKa = 4.02SYY124 pKa = 10.28VTEE127 pKa = 4.98DD128 pKa = 4.84EE129 pKa = 4.2FTEE132 pKa = 4.97DD133 pKa = 1.97IWGASRR139 pKa = 11.84NDD141 pKa = 4.18GIVYY145 pKa = 8.9GLPVDD150 pKa = 4.31CNPILMYY157 pKa = 10.51YY158 pKa = 10.42NKK160 pKa = 10.76ALLEE164 pKa = 4.28EE165 pKa = 4.48NNIPSPQEE173 pKa = 3.56LYY175 pKa = 11.21EE176 pKa = 4.26NGEE179 pKa = 4.4WNWDD183 pKa = 3.47TFEE186 pKa = 6.91DD187 pKa = 3.55ITGQLNEE194 pKa = 4.7SGNHH198 pKa = 5.96GYY200 pKa = 7.92MQEE203 pKa = 4.17SGHH206 pKa = 5.99GAMLNWIWTNGGEE219 pKa = 4.67FATEE223 pKa = 3.73DD224 pKa = 4.45AIVGDD229 pKa = 4.06TDD231 pKa = 3.69GKK233 pKa = 8.98TMEE236 pKa = 4.24AFEE239 pKa = 4.32YY240 pKa = 8.64VDD242 pKa = 4.56RR243 pKa = 11.84MIDD246 pKa = 3.38EE247 pKa = 5.1GNFTYY252 pKa = 10.6AGTMPEE258 pKa = 4.21GQGIEE263 pKa = 4.09AMFMSNQVGFVSAGRR278 pKa = 11.84WLTPMFLDD286 pKa = 3.71TSIDD290 pKa = 3.24FDD292 pKa = 4.79YY293 pKa = 11.26IPFPSNTDD301 pKa = 3.43EE302 pKa = 4.7QIEE305 pKa = 4.28TANIATAYY313 pKa = 8.55MAVNSKK319 pKa = 10.44SDD321 pKa = 3.46HH322 pKa = 6.46VEE324 pKa = 3.46EE325 pKa = 4.04AMKK328 pKa = 10.54FISYY332 pKa = 7.59YY333 pKa = 10.19TSTDD337 pKa = 3.32GQEE340 pKa = 3.6VRR342 pKa = 11.84LDD344 pKa = 3.76GEE346 pKa = 4.75GNAVPSVVGIDD357 pKa = 3.83EE358 pKa = 4.15IVLNQDD364 pKa = 3.18RR365 pKa = 11.84PEE367 pKa = 4.52HH368 pKa = 5.43GQYY371 pKa = 11.25LLDD374 pKa = 3.88ARR376 pKa = 11.84SVGRR380 pKa = 11.84VIGLEE385 pKa = 3.98FTSPGLSEE393 pKa = 4.7DD394 pKa = 4.22LEE396 pKa = 4.95DD397 pKa = 4.29IYY399 pKa = 11.34EE400 pKa = 4.25LMMLGDD406 pKa = 4.16IDD408 pKa = 5.54AGDD411 pKa = 4.32ALEE414 pKa = 4.19QATAKK419 pKa = 10.65AKK421 pKa = 10.61AAVEE425 pKa = 4.11
Molecular weight: 46.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A060M7N8|A0A060M7N8_9BACI Oligopeptide ABC transporter permease OS=Bacillus lehensis G1 OX=1246626 GN=BleG1_4024 PE=3 SV=1
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.16KK15 pKa = 9.1NHH17 pKa = 4.73GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.25NGRR29 pKa = 11.84KK30 pKa = 8.73VLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84QKK38 pKa = 10.43GRR40 pKa = 11.84KK41 pKa = 8.52VLSAA45 pKa = 4.05
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.16KK15 pKa = 9.1NHH17 pKa = 4.73GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.25NGRR29 pKa = 11.84KK30 pKa = 8.73VLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84QKK38 pKa = 10.43GRR40 pKa = 11.84KK41 pKa = 8.52VLSAA45 pKa = 4.05
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1166809 |
29 |
3427 |
290.5 |
32.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.492 ± 0.047 | 0.688 ± 0.011 |
5.153 ± 0.04 | 7.492 ± 0.045 |
4.537 ± 0.035 | 6.886 ± 0.049 |
2.368 ± 0.022 | 7.109 ± 0.04 |
5.766 ± 0.043 | 10.044 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.728 ± 0.021 | 3.954 ± 0.032 |
3.608 ± 0.023 | 4.138 ± 0.029 |
4.25 ± 0.028 | 6.115 ± 0.03 |
5.943 ± 0.042 | 7.219 ± 0.039 |
1.053 ± 0.014 | 3.456 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
