
Beihai rhabdo-like virus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Nyamiviridae; Berhavirus; Beihai berhavirus
Average proteome isoelectric point is 7.79
Get precalculated fractions of proteins
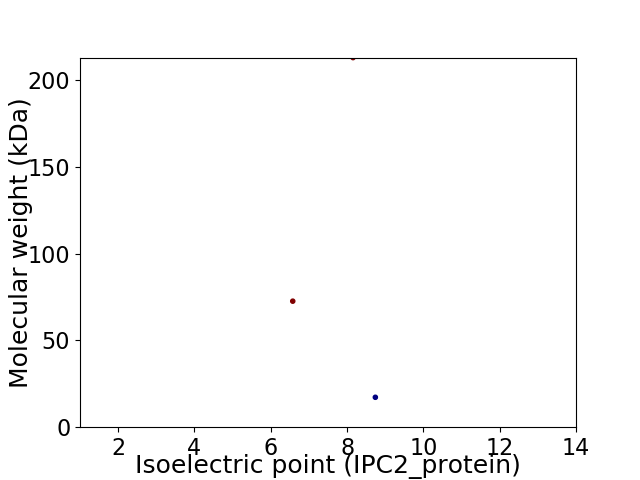
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KMN2|A0A1L3KMN2_9MONO Large structural protein OS=Beihai rhabdo-like virus 4 OX=1922654 PE=4 SV=1
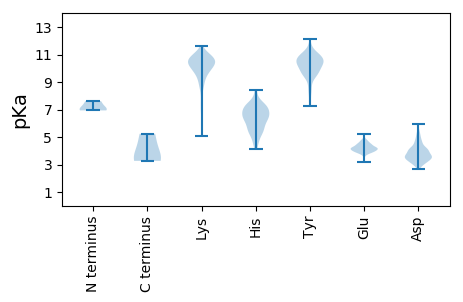
MM1 pKa = 7.03LQEE4 pKa = 4.45AGIEE8 pKa = 4.49FILMPTKK15 pKa = 10.56VFASGKK21 pKa = 10.29VNLKK25 pKa = 10.53DD26 pKa = 3.57QMSKK30 pKa = 10.97GLFWEE35 pKa = 4.7DD36 pKa = 4.17LGSLDD41 pKa = 4.56LLPYY45 pKa = 10.2ISNPRR50 pKa = 11.84VFNAVTEE57 pKa = 4.23LPEE60 pKa = 3.97TLKK63 pKa = 11.16YY64 pKa = 9.59CINMAGGINYY74 pKa = 9.55LGWTKK79 pKa = 10.72EE80 pKa = 4.13SAPGDD85 pKa = 3.98TPMSASSISTPAFPKK100 pKa = 8.7QTPAGGQGEE109 pKa = 4.71DD110 pKa = 3.99LLSAIQRR117 pKa = 11.84AAAEE121 pKa = 4.07RR122 pKa = 11.84AAQLQAEE129 pKa = 4.47EE130 pKa = 4.4EE131 pKa = 4.1ARR133 pKa = 11.84SRR135 pKa = 11.84SEE137 pKa = 4.03LRR139 pKa = 11.84QAQDD143 pKa = 2.64QDD145 pKa = 3.27GRR147 pKa = 11.84RR148 pKa = 11.84NPVADD153 pKa = 4.05RR154 pKa = 11.84DD155 pKa = 3.83HH156 pKa = 7.09RR157 pKa = 11.84PAGGGGGPGSSDD169 pKa = 3.24EE170 pKa = 4.22GDD172 pKa = 3.35EE173 pKa = 4.3TGDD176 pKa = 3.53DD177 pKa = 3.66VSSIEE182 pKa = 3.95SDD184 pKa = 3.55QEE186 pKa = 4.02GGEE189 pKa = 3.99VDD191 pKa = 3.58GQGTRR196 pKa = 11.84GGGAGGSTGRR206 pKa = 11.84GGGRR210 pKa = 11.84GDD212 pKa = 3.97NRR214 pKa = 11.84GTGQGEE220 pKa = 4.15GRR222 pKa = 11.84DD223 pKa = 3.53EE224 pKa = 4.03RR225 pKa = 11.84EE226 pKa = 3.52RR227 pKa = 11.84RR228 pKa = 11.84DD229 pKa = 3.27RR230 pKa = 11.84RR231 pKa = 11.84NRR233 pKa = 11.84RR234 pKa = 11.84TGRR237 pKa = 11.84GGRR240 pKa = 11.84DD241 pKa = 2.83RR242 pKa = 11.84RR243 pKa = 11.84GGRR246 pKa = 11.84GGRR249 pKa = 11.84GGGQDD254 pKa = 3.01RR255 pKa = 11.84DD256 pKa = 3.16RR257 pKa = 11.84DD258 pKa = 3.57RR259 pKa = 11.84RR260 pKa = 11.84EE261 pKa = 3.26ISGPQKK267 pKa = 10.64NKK269 pKa = 9.81VPRR272 pKa = 11.84EE273 pKa = 3.93MDD275 pKa = 3.37SAALGHH281 pKa = 6.77EE282 pKa = 4.4YY283 pKa = 10.42PEE285 pKa = 4.23ATLYY289 pKa = 10.2TDD291 pKa = 4.02PDD293 pKa = 3.22RR294 pKa = 11.84AYY296 pKa = 10.85ADD298 pKa = 3.71YY299 pKa = 10.82QRR301 pKa = 11.84EE302 pKa = 3.88ADD304 pKa = 4.16ALKK307 pKa = 9.98VWTPASEE314 pKa = 4.5VPRR317 pKa = 11.84PNPGLALINLLITCAWFTFNSLQIQNVRR345 pKa = 11.84AVRR348 pKa = 11.84SQPLKK353 pKa = 10.55PEE355 pKa = 4.21RR356 pKa = 11.84KK357 pKa = 7.42TDD359 pKa = 2.99AMRR362 pKa = 11.84NDD364 pKa = 3.2PHH366 pKa = 8.38LDD368 pKa = 3.15LRR370 pKa = 11.84YY371 pKa = 9.76LYY373 pKa = 10.56HH374 pKa = 6.96GFLAILCAIKK384 pKa = 10.63PMTEE388 pKa = 4.12DD389 pKa = 2.96NASYY393 pKa = 9.9VWKK396 pKa = 10.29RR397 pKa = 11.84FCAAVNSQDD406 pKa = 5.32LDD408 pKa = 3.54IEE410 pKa = 4.51LKK412 pKa = 10.96NKK414 pKa = 10.53LEE416 pKa = 4.02LVGVSLKK423 pKa = 10.53SLCDD427 pKa = 3.18AAAHH431 pKa = 6.23IKK433 pKa = 10.33RR434 pKa = 11.84YY435 pKa = 9.56IDD437 pKa = 3.81PKK439 pKa = 11.08DD440 pKa = 3.51VLSLLRR446 pKa = 11.84EE447 pKa = 4.14LDD449 pKa = 3.32VDD451 pKa = 4.85DD452 pKa = 5.92PDD454 pKa = 4.63LVAEE458 pKa = 4.7FDD460 pKa = 4.24LKK462 pKa = 10.68PVHH465 pKa = 6.98KK466 pKa = 10.63GLLQQMRR473 pKa = 11.84LIYY476 pKa = 10.54SGVAMARR483 pKa = 11.84IMQMEE488 pKa = 4.82KK489 pKa = 9.35WAQLFDD495 pKa = 4.64CRR497 pKa = 11.84AHH499 pKa = 5.64YY500 pKa = 10.59CNIVVSEE507 pKa = 4.55IISFLSIVKK516 pKa = 10.28ALRR519 pKa = 11.84KK520 pKa = 10.07KK521 pKa = 10.71FGEE524 pKa = 4.2DD525 pKa = 3.49FNFLRR530 pKa = 11.84IDD532 pKa = 3.55RR533 pKa = 11.84PKK535 pKa = 10.4EE536 pKa = 3.85LEE538 pKa = 4.03PLSSRR543 pKa = 11.84KK544 pKa = 9.02FPHH547 pKa = 6.55LAYY550 pKa = 10.44CATEE554 pKa = 3.81SGYY557 pKa = 7.27MTKK560 pKa = 10.53SFTRR564 pKa = 11.84KK565 pKa = 7.8MAKK568 pKa = 9.57SDD570 pKa = 3.75QPLMMPAKK578 pKa = 9.1EE579 pKa = 4.21LKK581 pKa = 10.95VMIKK585 pKa = 10.51DD586 pKa = 3.52RR587 pKa = 11.84LRR589 pKa = 11.84EE590 pKa = 3.92RR591 pKa = 11.84NILSPKK597 pKa = 9.92QILALRR603 pKa = 11.84SLGIALPEE611 pKa = 5.2EE612 pKa = 4.17KK613 pKa = 9.5TQWQGTKK620 pKa = 10.06RR621 pKa = 11.84PRR623 pKa = 11.84EE624 pKa = 4.14DD625 pKa = 3.08QAGPSTKK632 pKa = 9.92RR633 pKa = 11.84PKK635 pKa = 10.09PSNAYY640 pKa = 8.08FTEE643 pKa = 4.33SSEE646 pKa = 4.23EE647 pKa = 4.0EE648 pKa = 4.02SDD650 pKa = 3.56EE651 pKa = 4.77DD652 pKa = 4.32LL653 pKa = 5.24
MM1 pKa = 7.03LQEE4 pKa = 4.45AGIEE8 pKa = 4.49FILMPTKK15 pKa = 10.56VFASGKK21 pKa = 10.29VNLKK25 pKa = 10.53DD26 pKa = 3.57QMSKK30 pKa = 10.97GLFWEE35 pKa = 4.7DD36 pKa = 4.17LGSLDD41 pKa = 4.56LLPYY45 pKa = 10.2ISNPRR50 pKa = 11.84VFNAVTEE57 pKa = 4.23LPEE60 pKa = 3.97TLKK63 pKa = 11.16YY64 pKa = 9.59CINMAGGINYY74 pKa = 9.55LGWTKK79 pKa = 10.72EE80 pKa = 4.13SAPGDD85 pKa = 3.98TPMSASSISTPAFPKK100 pKa = 8.7QTPAGGQGEE109 pKa = 4.71DD110 pKa = 3.99LLSAIQRR117 pKa = 11.84AAAEE121 pKa = 4.07RR122 pKa = 11.84AAQLQAEE129 pKa = 4.47EE130 pKa = 4.4EE131 pKa = 4.1ARR133 pKa = 11.84SRR135 pKa = 11.84SEE137 pKa = 4.03LRR139 pKa = 11.84QAQDD143 pKa = 2.64QDD145 pKa = 3.27GRR147 pKa = 11.84RR148 pKa = 11.84NPVADD153 pKa = 4.05RR154 pKa = 11.84DD155 pKa = 3.83HH156 pKa = 7.09RR157 pKa = 11.84PAGGGGGPGSSDD169 pKa = 3.24EE170 pKa = 4.22GDD172 pKa = 3.35EE173 pKa = 4.3TGDD176 pKa = 3.53DD177 pKa = 3.66VSSIEE182 pKa = 3.95SDD184 pKa = 3.55QEE186 pKa = 4.02GGEE189 pKa = 3.99VDD191 pKa = 3.58GQGTRR196 pKa = 11.84GGGAGGSTGRR206 pKa = 11.84GGGRR210 pKa = 11.84GDD212 pKa = 3.97NRR214 pKa = 11.84GTGQGEE220 pKa = 4.15GRR222 pKa = 11.84DD223 pKa = 3.53EE224 pKa = 4.03RR225 pKa = 11.84EE226 pKa = 3.52RR227 pKa = 11.84RR228 pKa = 11.84DD229 pKa = 3.27RR230 pKa = 11.84RR231 pKa = 11.84NRR233 pKa = 11.84RR234 pKa = 11.84TGRR237 pKa = 11.84GGRR240 pKa = 11.84DD241 pKa = 2.83RR242 pKa = 11.84RR243 pKa = 11.84GGRR246 pKa = 11.84GGRR249 pKa = 11.84GGGQDD254 pKa = 3.01RR255 pKa = 11.84DD256 pKa = 3.16RR257 pKa = 11.84DD258 pKa = 3.57RR259 pKa = 11.84RR260 pKa = 11.84EE261 pKa = 3.26ISGPQKK267 pKa = 10.64NKK269 pKa = 9.81VPRR272 pKa = 11.84EE273 pKa = 3.93MDD275 pKa = 3.37SAALGHH281 pKa = 6.77EE282 pKa = 4.4YY283 pKa = 10.42PEE285 pKa = 4.23ATLYY289 pKa = 10.2TDD291 pKa = 4.02PDD293 pKa = 3.22RR294 pKa = 11.84AYY296 pKa = 10.85ADD298 pKa = 3.71YY299 pKa = 10.82QRR301 pKa = 11.84EE302 pKa = 3.88ADD304 pKa = 4.16ALKK307 pKa = 9.98VWTPASEE314 pKa = 4.5VPRR317 pKa = 11.84PNPGLALINLLITCAWFTFNSLQIQNVRR345 pKa = 11.84AVRR348 pKa = 11.84SQPLKK353 pKa = 10.55PEE355 pKa = 4.21RR356 pKa = 11.84KK357 pKa = 7.42TDD359 pKa = 2.99AMRR362 pKa = 11.84NDD364 pKa = 3.2PHH366 pKa = 8.38LDD368 pKa = 3.15LRR370 pKa = 11.84YY371 pKa = 9.76LYY373 pKa = 10.56HH374 pKa = 6.96GFLAILCAIKK384 pKa = 10.63PMTEE388 pKa = 4.12DD389 pKa = 2.96NASYY393 pKa = 9.9VWKK396 pKa = 10.29RR397 pKa = 11.84FCAAVNSQDD406 pKa = 5.32LDD408 pKa = 3.54IEE410 pKa = 4.51LKK412 pKa = 10.96NKK414 pKa = 10.53LEE416 pKa = 4.02LVGVSLKK423 pKa = 10.53SLCDD427 pKa = 3.18AAAHH431 pKa = 6.23IKK433 pKa = 10.33RR434 pKa = 11.84YY435 pKa = 9.56IDD437 pKa = 3.81PKK439 pKa = 11.08DD440 pKa = 3.51VLSLLRR446 pKa = 11.84EE447 pKa = 4.14LDD449 pKa = 3.32VDD451 pKa = 4.85DD452 pKa = 5.92PDD454 pKa = 4.63LVAEE458 pKa = 4.7FDD460 pKa = 4.24LKK462 pKa = 10.68PVHH465 pKa = 6.98KK466 pKa = 10.63GLLQQMRR473 pKa = 11.84LIYY476 pKa = 10.54SGVAMARR483 pKa = 11.84IMQMEE488 pKa = 4.82KK489 pKa = 9.35WAQLFDD495 pKa = 4.64CRR497 pKa = 11.84AHH499 pKa = 5.64YY500 pKa = 10.59CNIVVSEE507 pKa = 4.55IISFLSIVKK516 pKa = 10.28ALRR519 pKa = 11.84KK520 pKa = 10.07KK521 pKa = 10.71FGEE524 pKa = 4.2DD525 pKa = 3.49FNFLRR530 pKa = 11.84IDD532 pKa = 3.55RR533 pKa = 11.84PKK535 pKa = 10.4EE536 pKa = 3.85LEE538 pKa = 4.03PLSSRR543 pKa = 11.84KK544 pKa = 9.02FPHH547 pKa = 6.55LAYY550 pKa = 10.44CATEE554 pKa = 3.81SGYY557 pKa = 7.27MTKK560 pKa = 10.53SFTRR564 pKa = 11.84KK565 pKa = 7.8MAKK568 pKa = 9.57SDD570 pKa = 3.75QPLMMPAKK578 pKa = 9.1EE579 pKa = 4.21LKK581 pKa = 10.95VMIKK585 pKa = 10.51DD586 pKa = 3.52RR587 pKa = 11.84LRR589 pKa = 11.84EE590 pKa = 3.92RR591 pKa = 11.84NILSPKK597 pKa = 9.92QILALRR603 pKa = 11.84SLGIALPEE611 pKa = 5.2EE612 pKa = 4.17KK613 pKa = 9.5TQWQGTKK620 pKa = 10.06RR621 pKa = 11.84PRR623 pKa = 11.84EE624 pKa = 4.14DD625 pKa = 3.08QAGPSTKK632 pKa = 9.92RR633 pKa = 11.84PKK635 pKa = 10.09PSNAYY640 pKa = 8.08FTEE643 pKa = 4.33SSEE646 pKa = 4.23EE647 pKa = 4.0EE648 pKa = 4.02SDD650 pKa = 3.56EE651 pKa = 4.77DD652 pKa = 4.32LL653 pKa = 5.24
Molecular weight: 72.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMQ1|A0A1L3KMQ1_9MONO Uncharacterized protein OS=Beihai rhabdo-like virus 4 OX=1922654 PE=4 SV=1
MM1 pKa = 7.61SGRR4 pKa = 11.84IRR6 pKa = 11.84PRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84LPQVSSAQAAGGAEE24 pKa = 4.06RR25 pKa = 11.84FRR27 pKa = 11.84CYY29 pKa = 10.52LGVTFRR35 pKa = 11.84FQNGLSLHH43 pKa = 5.29TVKK46 pKa = 11.08SNILAIHH53 pKa = 6.84LKK55 pKa = 9.97RR56 pKa = 11.84PYY58 pKa = 10.23VSRR61 pKa = 11.84GYY63 pKa = 10.01IRR65 pKa = 11.84EE66 pKa = 4.21LKK68 pKa = 9.7LTLQRR73 pKa = 11.84VGGPSRR79 pKa = 11.84IAVRR83 pKa = 11.84QPSGDD88 pKa = 3.57NPQTPTVCHH97 pKa = 5.43VCRR100 pKa = 11.84DD101 pKa = 3.43EE102 pKa = 4.75RR103 pKa = 11.84PIYY106 pKa = 9.63TYY108 pKa = 10.84LWSDD112 pKa = 3.51LGISEE117 pKa = 4.49TWCKK121 pKa = 10.66GDD123 pKa = 3.52EE124 pKa = 4.24APTISVDD131 pKa = 3.14NFKK134 pKa = 10.7VARR137 pKa = 11.84VLEE140 pKa = 4.49DD141 pKa = 4.49GYY143 pKa = 11.58CEE145 pKa = 3.89FLYY148 pKa = 10.71TEE150 pKa = 4.44TCC152 pKa = 3.47
MM1 pKa = 7.61SGRR4 pKa = 11.84IRR6 pKa = 11.84PRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84LPQVSSAQAAGGAEE24 pKa = 4.06RR25 pKa = 11.84FRR27 pKa = 11.84CYY29 pKa = 10.52LGVTFRR35 pKa = 11.84FQNGLSLHH43 pKa = 5.29TVKK46 pKa = 11.08SNILAIHH53 pKa = 6.84LKK55 pKa = 9.97RR56 pKa = 11.84PYY58 pKa = 10.23VSRR61 pKa = 11.84GYY63 pKa = 10.01IRR65 pKa = 11.84EE66 pKa = 4.21LKK68 pKa = 9.7LTLQRR73 pKa = 11.84VGGPSRR79 pKa = 11.84IAVRR83 pKa = 11.84QPSGDD88 pKa = 3.57NPQTPTVCHH97 pKa = 5.43VCRR100 pKa = 11.84DD101 pKa = 3.43EE102 pKa = 4.75RR103 pKa = 11.84PIYY106 pKa = 9.63TYY108 pKa = 10.84LWSDD112 pKa = 3.51LGISEE117 pKa = 4.49TWCKK121 pKa = 10.66GDD123 pKa = 3.52EE124 pKa = 4.24APTISVDD131 pKa = 3.14NFKK134 pKa = 10.7VARR137 pKa = 11.84VLEE140 pKa = 4.49DD141 pKa = 4.49GYY143 pKa = 11.58CEE145 pKa = 3.89FLYY148 pKa = 10.71TEE150 pKa = 4.44TCC152 pKa = 3.47
Molecular weight: 17.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2661 |
152 |
1856 |
887.0 |
100.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.013 ± 1.0 | 2.067 ± 0.611 |
6.201 ± 0.806 | 5.787 ± 0.693 |
4.247 ± 0.747 | 6.276 ± 1.429 |
2.518 ± 0.614 | 5.637 ± 0.623 |
6.464 ± 0.977 | 9.583 ± 0.359 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.818 ± 0.631 | 3.983 ± 0.544 |
4.434 ± 0.789 | 3.457 ± 0.476 |
7.253 ± 1.446 | 6.99 ± 0.124 |
5.224 ± 0.632 | 6.276 ± 1.089 |
1.24 ± 0.075 | 3.533 ± 0.536 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
