
Influenza D virus (D/bovine/France/2986/2012)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Insthoviricetes; Articulavirales; Orthomyxoviridae; Deltainfluenzavirus; Influenza D virus
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
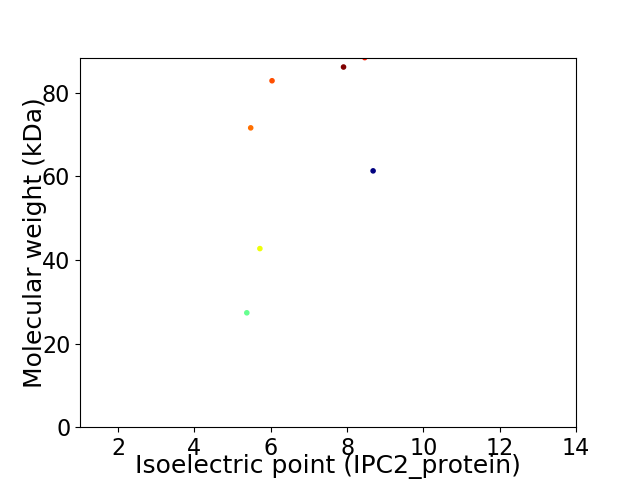
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E4C2T3|A0A0E4C2T3_9ORTO p3 OS=Influenza D virus (D/bovine/France/2986/2012) OX=1542973 GN=P3 PE=4 SV=1
MM1 pKa = 7.43FLLLATITAITACQAEE17 pKa = 4.47RR18 pKa = 11.84EE19 pKa = 4.52LICIVQRR26 pKa = 11.84VNEE29 pKa = 4.38SFSLHH34 pKa = 6.32SGFGGNVYY42 pKa = 10.76SMKK45 pKa = 9.78TEE47 pKa = 3.84PMTGFTNVTKK57 pKa = 10.27GASVINQKK65 pKa = 10.62DD66 pKa = 3.32WIGFGDD72 pKa = 4.17SRR74 pKa = 11.84TDD76 pKa = 3.16LTNAQFPASSDD87 pKa = 3.52VPLAVAKK94 pKa = 10.39KK95 pKa = 9.46FRR97 pKa = 11.84SLSGASLMLSAFGPPDD113 pKa = 3.46KK114 pKa = 10.91VDD116 pKa = 3.68YY117 pKa = 10.65LYY119 pKa = 10.71QGCGKK124 pKa = 10.19EE125 pKa = 3.91KK126 pKa = 10.66VFYY129 pKa = 10.74EE130 pKa = 4.47GVNWSPEE137 pKa = 3.71AGIDD141 pKa = 3.84CFGSNWTQTKK151 pKa = 10.06KK152 pKa = 10.77DD153 pKa = 4.01FYY155 pKa = 11.06SRR157 pKa = 11.84IYY159 pKa = 9.57EE160 pKa = 3.98AARR163 pKa = 11.84GSTCMTLVNSLDD175 pKa = 3.78TKK177 pKa = 10.63ISSTTATAGTTSSCSSSWMKK197 pKa = 10.82SPLWYY202 pKa = 10.36AEE204 pKa = 3.96SSVNPGASPQVCGTEE219 pKa = 3.81QSAIFTLPTSFGIYY233 pKa = 9.38KK234 pKa = 9.96CNKK237 pKa = 9.4HH238 pKa = 6.03VVQLCYY244 pKa = 9.81FVYY247 pKa = 10.38EE248 pKa = 4.55SKK250 pKa = 9.54TAFNTFGCGDD260 pKa = 4.19YY261 pKa = 10.37YY262 pKa = 11.22QNYY265 pKa = 9.53YY266 pKa = 10.46DD267 pKa = 4.84GNGNLIGGMDD277 pKa = 3.91NRR279 pKa = 11.84VAAYY283 pKa = 10.04RR284 pKa = 11.84GIAGAGAKK292 pKa = 9.46IEE294 pKa = 4.41CPSKK298 pKa = 10.3ILNPGTYY305 pKa = 9.9SIRR308 pKa = 11.84STPRR312 pKa = 11.84FLLVPKK318 pKa = 10.37RR319 pKa = 11.84SYY321 pKa = 11.48CFDD324 pKa = 3.44TDD326 pKa = 2.79GGYY329 pKa = 9.81PIQVVQSEE337 pKa = 4.36WSASRR342 pKa = 11.84RR343 pKa = 11.84SDD345 pKa = 3.09NATEE349 pKa = 4.1EE350 pKa = 4.53ACLQTEE356 pKa = 4.29GCIFIKK362 pKa = 9.79KK363 pKa = 4.33TTPYY367 pKa = 10.51VGEE370 pKa = 4.65ADD372 pKa = 4.66DD373 pKa = 3.73NHH375 pKa = 8.17GDD377 pKa = 3.19IEE379 pKa = 4.21MRR381 pKa = 11.84QLLSGLGNNDD391 pKa = 3.69TVCVSQSGYY400 pKa = 9.38TKK402 pKa = 11.12GEE404 pKa = 4.05TPFVKK409 pKa = 10.25DD410 pKa = 3.32YY411 pKa = 11.12LSPPKK416 pKa = 10.44YY417 pKa = 10.38GRR419 pKa = 11.84CQLKK423 pKa = 9.04TDD425 pKa = 3.58SGRR428 pKa = 11.84IPTLPSGLIIPQAGTDD444 pKa = 3.32SLMRR448 pKa = 11.84TLTPATRR455 pKa = 11.84IFGIDD460 pKa = 3.46DD461 pKa = 5.14LIFGLLFIGFVAGGVAGGYY480 pKa = 8.51FWGRR484 pKa = 11.84SSEE487 pKa = 4.23GGGGASVSSTQAGFDD502 pKa = 4.2KK503 pKa = 10.45IGKK506 pKa = 9.3DD507 pKa = 3.34IQQLRR512 pKa = 11.84SDD514 pKa = 3.68TNAAIEE520 pKa = 4.27GFNGRR525 pKa = 11.84IAHH528 pKa = 7.12DD529 pKa = 3.64EE530 pKa = 3.99QAVKK534 pKa = 10.83NLAKK538 pKa = 10.26EE539 pKa = 4.12IEE541 pKa = 4.3DD542 pKa = 3.64ARR544 pKa = 11.84AEE546 pKa = 4.09ALVGEE551 pKa = 4.44LGIIRR556 pKa = 11.84SLIVANISMNLKK568 pKa = 9.73EE569 pKa = 4.66SLYY572 pKa = 10.74EE573 pKa = 3.92LANQITKK580 pKa = 10.42RR581 pKa = 11.84GGGIAQEE588 pKa = 4.44AGPGCWYY595 pKa = 10.6VDD597 pKa = 4.53SEE599 pKa = 4.67NCDD602 pKa = 3.46ASCKK606 pKa = 10.28EE607 pKa = 4.07YY608 pKa = 10.57IFNFNGSATVPTLRR622 pKa = 11.84PVDD625 pKa = 3.66TNVIITSDD633 pKa = 3.75PYY635 pKa = 11.64YY636 pKa = 10.56LGSTIALCLLGLVAIAAFVGVIWICCKK663 pKa = 10.45KK664 pKa = 10.73
MM1 pKa = 7.43FLLLATITAITACQAEE17 pKa = 4.47RR18 pKa = 11.84EE19 pKa = 4.52LICIVQRR26 pKa = 11.84VNEE29 pKa = 4.38SFSLHH34 pKa = 6.32SGFGGNVYY42 pKa = 10.76SMKK45 pKa = 9.78TEE47 pKa = 3.84PMTGFTNVTKK57 pKa = 10.27GASVINQKK65 pKa = 10.62DD66 pKa = 3.32WIGFGDD72 pKa = 4.17SRR74 pKa = 11.84TDD76 pKa = 3.16LTNAQFPASSDD87 pKa = 3.52VPLAVAKK94 pKa = 10.39KK95 pKa = 9.46FRR97 pKa = 11.84SLSGASLMLSAFGPPDD113 pKa = 3.46KK114 pKa = 10.91VDD116 pKa = 3.68YY117 pKa = 10.65LYY119 pKa = 10.71QGCGKK124 pKa = 10.19EE125 pKa = 3.91KK126 pKa = 10.66VFYY129 pKa = 10.74EE130 pKa = 4.47GVNWSPEE137 pKa = 3.71AGIDD141 pKa = 3.84CFGSNWTQTKK151 pKa = 10.06KK152 pKa = 10.77DD153 pKa = 4.01FYY155 pKa = 11.06SRR157 pKa = 11.84IYY159 pKa = 9.57EE160 pKa = 3.98AARR163 pKa = 11.84GSTCMTLVNSLDD175 pKa = 3.78TKK177 pKa = 10.63ISSTTATAGTTSSCSSSWMKK197 pKa = 10.82SPLWYY202 pKa = 10.36AEE204 pKa = 3.96SSVNPGASPQVCGTEE219 pKa = 3.81QSAIFTLPTSFGIYY233 pKa = 9.38KK234 pKa = 9.96CNKK237 pKa = 9.4HH238 pKa = 6.03VVQLCYY244 pKa = 9.81FVYY247 pKa = 10.38EE248 pKa = 4.55SKK250 pKa = 9.54TAFNTFGCGDD260 pKa = 4.19YY261 pKa = 10.37YY262 pKa = 11.22QNYY265 pKa = 9.53YY266 pKa = 10.46DD267 pKa = 4.84GNGNLIGGMDD277 pKa = 3.91NRR279 pKa = 11.84VAAYY283 pKa = 10.04RR284 pKa = 11.84GIAGAGAKK292 pKa = 9.46IEE294 pKa = 4.41CPSKK298 pKa = 10.3ILNPGTYY305 pKa = 9.9SIRR308 pKa = 11.84STPRR312 pKa = 11.84FLLVPKK318 pKa = 10.37RR319 pKa = 11.84SYY321 pKa = 11.48CFDD324 pKa = 3.44TDD326 pKa = 2.79GGYY329 pKa = 9.81PIQVVQSEE337 pKa = 4.36WSASRR342 pKa = 11.84RR343 pKa = 11.84SDD345 pKa = 3.09NATEE349 pKa = 4.1EE350 pKa = 4.53ACLQTEE356 pKa = 4.29GCIFIKK362 pKa = 9.79KK363 pKa = 4.33TTPYY367 pKa = 10.51VGEE370 pKa = 4.65ADD372 pKa = 4.66DD373 pKa = 3.73NHH375 pKa = 8.17GDD377 pKa = 3.19IEE379 pKa = 4.21MRR381 pKa = 11.84QLLSGLGNNDD391 pKa = 3.69TVCVSQSGYY400 pKa = 9.38TKK402 pKa = 11.12GEE404 pKa = 4.05TPFVKK409 pKa = 10.25DD410 pKa = 3.32YY411 pKa = 11.12LSPPKK416 pKa = 10.44YY417 pKa = 10.38GRR419 pKa = 11.84CQLKK423 pKa = 9.04TDD425 pKa = 3.58SGRR428 pKa = 11.84IPTLPSGLIIPQAGTDD444 pKa = 3.32SLMRR448 pKa = 11.84TLTPATRR455 pKa = 11.84IFGIDD460 pKa = 3.46DD461 pKa = 5.14LIFGLLFIGFVAGGVAGGYY480 pKa = 8.51FWGRR484 pKa = 11.84SSEE487 pKa = 4.23GGGGASVSSTQAGFDD502 pKa = 4.2KK503 pKa = 10.45IGKK506 pKa = 9.3DD507 pKa = 3.34IQQLRR512 pKa = 11.84SDD514 pKa = 3.68TNAAIEE520 pKa = 4.27GFNGRR525 pKa = 11.84IAHH528 pKa = 7.12DD529 pKa = 3.64EE530 pKa = 3.99QAVKK534 pKa = 10.83NLAKK538 pKa = 10.26EE539 pKa = 4.12IEE541 pKa = 4.3DD542 pKa = 3.64ARR544 pKa = 11.84AEE546 pKa = 4.09ALVGEE551 pKa = 4.44LGIIRR556 pKa = 11.84SLIVANISMNLKK568 pKa = 9.73EE569 pKa = 4.66SLYY572 pKa = 10.74EE573 pKa = 3.92LANQITKK580 pKa = 10.42RR581 pKa = 11.84GGGIAQEE588 pKa = 4.44AGPGCWYY595 pKa = 10.6VDD597 pKa = 4.53SEE599 pKa = 4.67NCDD602 pKa = 3.46ASCKK606 pKa = 10.28EE607 pKa = 4.07YY608 pKa = 10.57IFNFNGSATVPTLRR622 pKa = 11.84PVDD625 pKa = 3.66TNVIITSDD633 pKa = 3.75PYY635 pKa = 11.64YY636 pKa = 10.56LGSTIALCLLGLVAIAAFVGVIWICCKK663 pKa = 10.45KK664 pKa = 10.73
Molecular weight: 71.59 kDa
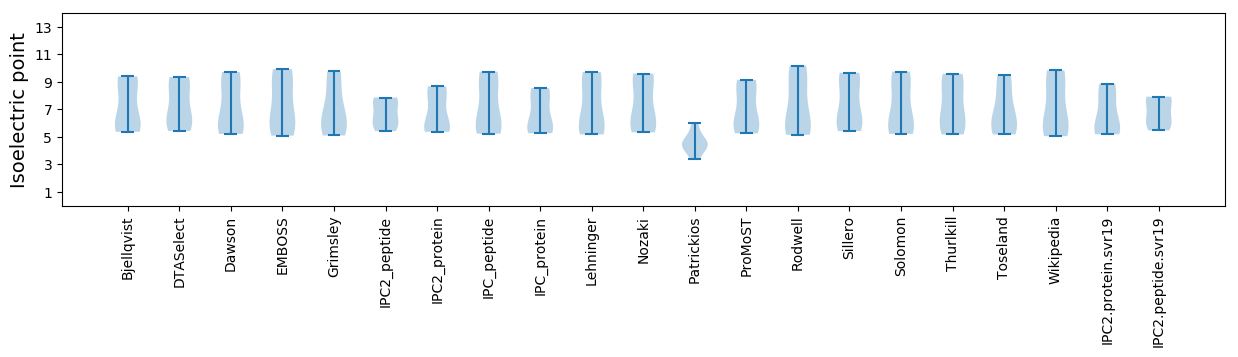
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E4C2S8|A0A0E4C2S8_9ORTO PB2 OS=Influenza D virus (D/bovine/France/2986/2012) OX=1542973 GN=PB2 PE=4 SV=1
MM1 pKa = 8.3DD2 pKa = 3.59STKK5 pKa = 10.9AQTPEE10 pKa = 3.5EE11 pKa = 3.96QRR13 pKa = 11.84AKK15 pKa = 9.92NAKK18 pKa = 9.12TILEE22 pKa = 4.4NIQIYY27 pKa = 10.6EE28 pKa = 4.19RR29 pKa = 11.84MCDD32 pKa = 3.34LFGVSEE38 pKa = 4.37DD39 pKa = 3.87DD40 pKa = 4.4KK41 pKa = 11.8LIIEE45 pKa = 4.25NSISIEE51 pKa = 3.75RR52 pKa = 11.84MIRR55 pKa = 11.84VVTDD59 pKa = 3.53KK60 pKa = 10.94KK61 pKa = 11.15YY62 pKa = 10.68QDD64 pKa = 3.89KK65 pKa = 10.62KK66 pKa = 10.92LKK68 pKa = 10.42NAGSDD73 pKa = 3.52LEE75 pKa = 4.69KK76 pKa = 10.33IANAGKK82 pKa = 9.29VFCRR86 pKa = 11.84LVEE89 pKa = 4.33STAGKK94 pKa = 10.19CSARR98 pKa = 11.84LGMALKK104 pKa = 10.48PNVEE108 pKa = 4.11AVLTDD113 pKa = 3.36VLGNEE118 pKa = 4.47LDD120 pKa = 3.58RR121 pKa = 11.84AAVLGKK127 pKa = 10.69RR128 pKa = 11.84MGFTAMFKK136 pKa = 10.97SNLEE140 pKa = 3.77EE141 pKa = 3.69VLYY144 pKa = 10.89QRR146 pKa = 11.84GKK148 pKa = 9.78NQLKK152 pKa = 9.94KK153 pKa = 10.37RR154 pKa = 11.84NSAEE158 pKa = 4.17TFTLSQGASLEE169 pKa = 4.05ARR171 pKa = 11.84FRR173 pKa = 11.84PIMEE177 pKa = 3.92KK178 pKa = 10.2HH179 pKa = 6.02LGVGTVVASIKK190 pKa = 10.84NILASKK196 pKa = 10.75KK197 pKa = 10.16NGNYY201 pKa = 8.42KK202 pKa = 10.65NKK204 pKa = 9.35MVRR207 pKa = 11.84KK208 pKa = 8.61PGGNRR213 pKa = 11.84EE214 pKa = 3.96SWSPLEE220 pKa = 4.41RR221 pKa = 11.84EE222 pKa = 3.84ISFLNKK228 pKa = 9.93KK229 pKa = 9.56LFPGPMRR236 pKa = 11.84QLCKK240 pKa = 10.45KK241 pKa = 10.55FEE243 pKa = 4.07YY244 pKa = 11.12LNDD247 pKa = 3.61QEE249 pKa = 5.2KK250 pKa = 10.37QLALNLMLDD259 pKa = 3.5ASLILKK265 pKa = 8.33PQVTHH270 pKa = 6.86KK271 pKa = 10.49MIMPWSMWLAVKK283 pKa = 10.24KK284 pKa = 9.74YY285 pKa = 10.78AEE287 pKa = 4.23MNKK290 pKa = 9.89GSPSLEE296 pKa = 3.86DD297 pKa = 3.12LAAYY301 pKa = 10.16SGVRR305 pKa = 11.84AFMAFNTACYY315 pKa = 9.03MSKK318 pKa = 9.15FTIGKK323 pKa = 9.63GIVGDD328 pKa = 3.95AEE330 pKa = 3.97IMEE333 pKa = 4.57NGNDD337 pKa = 3.51KK338 pKa = 9.86MQILAMACFGLAYY351 pKa = 9.73EE352 pKa = 4.54DD353 pKa = 3.77TGIVAAMISQPMKK366 pKa = 10.25KK367 pKa = 9.71RR368 pKa = 11.84YY369 pKa = 7.14QLKK372 pKa = 9.76VGNFNPPEE380 pKa = 4.11EE381 pKa = 4.33GTIKK385 pKa = 9.77GTSAGYY391 pKa = 6.91FHH393 pKa = 7.42KK394 pKa = 10.11WAEE397 pKa = 4.05FGNRR401 pKa = 11.84LPFNSFGTGEE411 pKa = 4.3SKK413 pKa = 10.73QISNSGVFAVQRR425 pKa = 11.84PSTTNIQRR433 pKa = 11.84LAEE436 pKa = 3.98LMARR440 pKa = 11.84NTGEE444 pKa = 3.77TSDD447 pKa = 4.85NFTQLVQKK455 pKa = 10.37IRR457 pKa = 11.84EE458 pKa = 4.18QVGTFADD465 pKa = 3.6QKK467 pKa = 10.78ANLRR471 pKa = 11.84EE472 pKa = 3.95FTGGYY477 pKa = 9.5IYY479 pKa = 10.36DD480 pKa = 3.59ITDD483 pKa = 3.38VTKK486 pKa = 10.73SNPKK490 pKa = 9.71IPQLGGNSFFFEE502 pKa = 4.45FTGSDD507 pKa = 3.7VPRR510 pKa = 11.84TGAKK514 pKa = 9.49RR515 pKa = 11.84RR516 pKa = 11.84VGGADD521 pKa = 3.6DD522 pKa = 3.83VTLGTSQPKK531 pKa = 9.75KK532 pKa = 9.96RR533 pKa = 11.84GRR535 pKa = 11.84QGAGVEE541 pKa = 3.94SSMDD545 pKa = 3.15IEE547 pKa = 4.89TVGEE551 pKa = 4.13DD552 pKa = 3.08
MM1 pKa = 8.3DD2 pKa = 3.59STKK5 pKa = 10.9AQTPEE10 pKa = 3.5EE11 pKa = 3.96QRR13 pKa = 11.84AKK15 pKa = 9.92NAKK18 pKa = 9.12TILEE22 pKa = 4.4NIQIYY27 pKa = 10.6EE28 pKa = 4.19RR29 pKa = 11.84MCDD32 pKa = 3.34LFGVSEE38 pKa = 4.37DD39 pKa = 3.87DD40 pKa = 4.4KK41 pKa = 11.8LIIEE45 pKa = 4.25NSISIEE51 pKa = 3.75RR52 pKa = 11.84MIRR55 pKa = 11.84VVTDD59 pKa = 3.53KK60 pKa = 10.94KK61 pKa = 11.15YY62 pKa = 10.68QDD64 pKa = 3.89KK65 pKa = 10.62KK66 pKa = 10.92LKK68 pKa = 10.42NAGSDD73 pKa = 3.52LEE75 pKa = 4.69KK76 pKa = 10.33IANAGKK82 pKa = 9.29VFCRR86 pKa = 11.84LVEE89 pKa = 4.33STAGKK94 pKa = 10.19CSARR98 pKa = 11.84LGMALKK104 pKa = 10.48PNVEE108 pKa = 4.11AVLTDD113 pKa = 3.36VLGNEE118 pKa = 4.47LDD120 pKa = 3.58RR121 pKa = 11.84AAVLGKK127 pKa = 10.69RR128 pKa = 11.84MGFTAMFKK136 pKa = 10.97SNLEE140 pKa = 3.77EE141 pKa = 3.69VLYY144 pKa = 10.89QRR146 pKa = 11.84GKK148 pKa = 9.78NQLKK152 pKa = 9.94KK153 pKa = 10.37RR154 pKa = 11.84NSAEE158 pKa = 4.17TFTLSQGASLEE169 pKa = 4.05ARR171 pKa = 11.84FRR173 pKa = 11.84PIMEE177 pKa = 3.92KK178 pKa = 10.2HH179 pKa = 6.02LGVGTVVASIKK190 pKa = 10.84NILASKK196 pKa = 10.75KK197 pKa = 10.16NGNYY201 pKa = 8.42KK202 pKa = 10.65NKK204 pKa = 9.35MVRR207 pKa = 11.84KK208 pKa = 8.61PGGNRR213 pKa = 11.84EE214 pKa = 3.96SWSPLEE220 pKa = 4.41RR221 pKa = 11.84EE222 pKa = 3.84ISFLNKK228 pKa = 9.93KK229 pKa = 9.56LFPGPMRR236 pKa = 11.84QLCKK240 pKa = 10.45KK241 pKa = 10.55FEE243 pKa = 4.07YY244 pKa = 11.12LNDD247 pKa = 3.61QEE249 pKa = 5.2KK250 pKa = 10.37QLALNLMLDD259 pKa = 3.5ASLILKK265 pKa = 8.33PQVTHH270 pKa = 6.86KK271 pKa = 10.49MIMPWSMWLAVKK283 pKa = 10.24KK284 pKa = 9.74YY285 pKa = 10.78AEE287 pKa = 4.23MNKK290 pKa = 9.89GSPSLEE296 pKa = 3.86DD297 pKa = 3.12LAAYY301 pKa = 10.16SGVRR305 pKa = 11.84AFMAFNTACYY315 pKa = 9.03MSKK318 pKa = 9.15FTIGKK323 pKa = 9.63GIVGDD328 pKa = 3.95AEE330 pKa = 3.97IMEE333 pKa = 4.57NGNDD337 pKa = 3.51KK338 pKa = 9.86MQILAMACFGLAYY351 pKa = 9.73EE352 pKa = 4.54DD353 pKa = 3.77TGIVAAMISQPMKK366 pKa = 10.25KK367 pKa = 9.71RR368 pKa = 11.84YY369 pKa = 7.14QLKK372 pKa = 9.76VGNFNPPEE380 pKa = 4.11EE381 pKa = 4.33GTIKK385 pKa = 9.77GTSAGYY391 pKa = 6.91FHH393 pKa = 7.42KK394 pKa = 10.11WAEE397 pKa = 4.05FGNRR401 pKa = 11.84LPFNSFGTGEE411 pKa = 4.3SKK413 pKa = 10.73QISNSGVFAVQRR425 pKa = 11.84PSTTNIQRR433 pKa = 11.84LAEE436 pKa = 3.98LMARR440 pKa = 11.84NTGEE444 pKa = 3.77TSDD447 pKa = 4.85NFTQLVQKK455 pKa = 10.37IRR457 pKa = 11.84EE458 pKa = 4.18QVGTFADD465 pKa = 3.6QKK467 pKa = 10.78ANLRR471 pKa = 11.84EE472 pKa = 3.95FTGGYY477 pKa = 9.5IYY479 pKa = 10.36DD480 pKa = 3.59ITDD483 pKa = 3.38VTKK486 pKa = 10.73SNPKK490 pKa = 9.71IPQLGGNSFFFEE502 pKa = 4.45FTGSDD507 pKa = 3.7VPRR510 pKa = 11.84TGAKK514 pKa = 9.49RR515 pKa = 11.84RR516 pKa = 11.84VGGADD521 pKa = 3.6DD522 pKa = 3.83VTLGTSQPKK531 pKa = 9.75KK532 pKa = 9.96RR533 pKa = 11.84GRR535 pKa = 11.84QGAGVEE541 pKa = 3.94SSMDD545 pKa = 3.15IEE547 pKa = 4.89TVGEE551 pKa = 4.13DD552 pKa = 3.08
Molecular weight: 61.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4081 |
243 |
772 |
583.0 |
65.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.641 ± 0.606 | 2.23 ± 0.27 |
4.631 ± 0.215 | 8.233 ± 0.805 |
4.631 ± 0.428 | 7.131 ± 0.726 |
1.054 ± 0.188 | 6.028 ± 0.209 |
8.013 ± 0.573 | 8.233 ± 0.388 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.994 ± 0.724 | 4.729 ± 0.309 |
3.651 ± 0.154 | 2.965 ± 0.262 |
5.587 ± 0.413 | 6.714 ± 0.459 |
5.954 ± 0.404 | 5.342 ± 0.388 |
1.201 ± 0.134 | 3.038 ± 0.232 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
