
Microbacterium phage Burro
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Burrovirus; Microbacterium virus Burro
Average proteome isoelectric point is 5.51
Get precalculated fractions of proteins
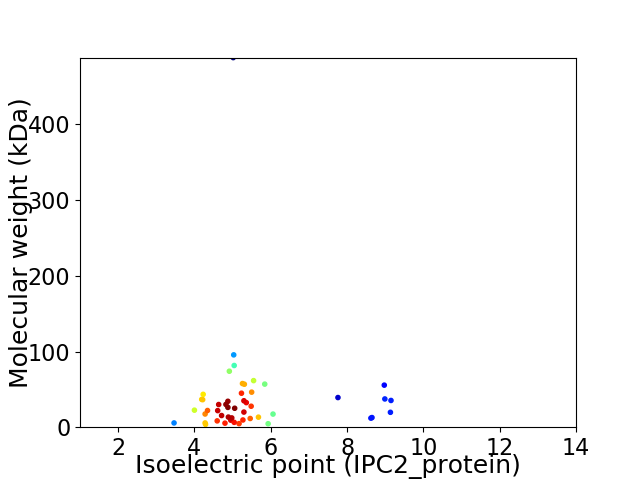
Virtual 2D-PAGE plot for 49 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
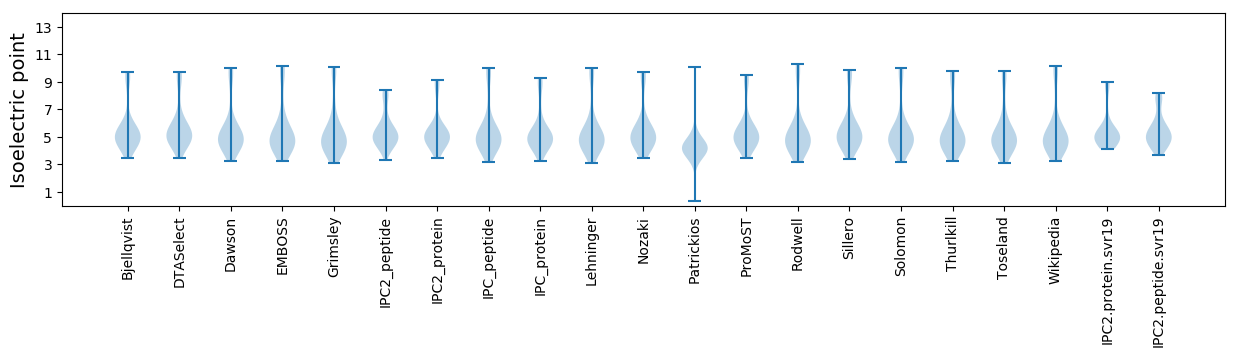
* You can choose from 21 different methods for calculating isoelectric point
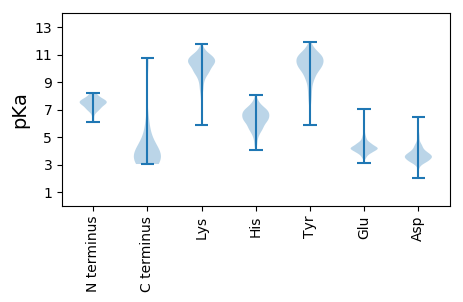
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A386KMN1|A0A386KMN1_9CAUD Uncharacterized protein OS=Microbacterium phage Burro OX=2315703 GN=46 PE=4 SV=1
MM1 pKa = 7.56SEE3 pKa = 3.76NDD5 pKa = 3.31EE6 pKa = 4.63TVVNTAIVDD15 pKa = 3.93TSPGAIVAATFEE27 pKa = 4.43KK28 pKa = 10.68PEE30 pKa = 4.01PLKK33 pKa = 10.84RR34 pKa = 11.84QWFLYY39 pKa = 9.84AAAAILLVATVILGSAFITNLTDD62 pKa = 4.0RR63 pKa = 11.84NNRR66 pKa = 11.84LNNVIVSQQDD76 pKa = 3.43SLHH79 pKa = 6.53EE80 pKa = 4.19KK81 pKa = 10.6DD82 pKa = 3.74EE83 pKa = 4.37QIEE86 pKa = 4.23EE87 pKa = 4.23LTATSQALYY96 pKa = 10.58DD97 pKa = 3.77QLLLAGEE104 pKa = 4.61TPDD107 pKa = 3.6EE108 pKa = 4.39PRR110 pKa = 11.84PPDD113 pKa = 3.92VVTGPAGDD121 pKa = 3.68RR122 pKa = 11.84GATGEE127 pKa = 4.24TGEE130 pKa = 4.61TGPPGPAGPVGPPGSQGEE148 pKa = 4.19PGVAGSSGNQGEE160 pKa = 4.35PGATGPSGPAGPAGPQGEE178 pKa = 4.5PGSPGATGPAGQSAFPFTFTFTYY201 pKa = 10.55LGIPYY206 pKa = 8.58TCTVTDD212 pKa = 3.5NTTAAACSITPP223 pKa = 3.7
MM1 pKa = 7.56SEE3 pKa = 3.76NDD5 pKa = 3.31EE6 pKa = 4.63TVVNTAIVDD15 pKa = 3.93TSPGAIVAATFEE27 pKa = 4.43KK28 pKa = 10.68PEE30 pKa = 4.01PLKK33 pKa = 10.84RR34 pKa = 11.84QWFLYY39 pKa = 9.84AAAAILLVATVILGSAFITNLTDD62 pKa = 4.0RR63 pKa = 11.84NNRR66 pKa = 11.84LNNVIVSQQDD76 pKa = 3.43SLHH79 pKa = 6.53EE80 pKa = 4.19KK81 pKa = 10.6DD82 pKa = 3.74EE83 pKa = 4.37QIEE86 pKa = 4.23EE87 pKa = 4.23LTATSQALYY96 pKa = 10.58DD97 pKa = 3.77QLLLAGEE104 pKa = 4.61TPDD107 pKa = 3.6EE108 pKa = 4.39PRR110 pKa = 11.84PPDD113 pKa = 3.92VVTGPAGDD121 pKa = 3.68RR122 pKa = 11.84GATGEE127 pKa = 4.24TGEE130 pKa = 4.61TGPPGPAGPVGPPGSQGEE148 pKa = 4.19PGVAGSSGNQGEE160 pKa = 4.35PGATGPSGPAGPAGPQGEE178 pKa = 4.5PGSPGATGPAGQSAFPFTFTFTYY201 pKa = 10.55LGIPYY206 pKa = 8.58TCTVTDD212 pKa = 3.5NTTAAACSITPP223 pKa = 3.7
Molecular weight: 22.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A386KL89|A0A386KL89_9CAUD Uncharacterized protein OS=Microbacterium phage Burro OX=2315703 GN=49 PE=4 SV=1
MM1 pKa = 7.65AKK3 pKa = 10.13KK4 pKa = 8.85KK5 pKa = 9.33TEE7 pKa = 4.38TIGEE11 pKa = 4.17RR12 pKa = 11.84NKK14 pKa = 10.79RR15 pKa = 11.84LGNAPGTGSVLKK27 pKa = 10.25PGYY30 pKa = 6.92TAPASPWGSLGNFLNPKK47 pKa = 8.77KK48 pKa = 10.03QVPVAPQNPRR58 pKa = 11.84TVTTAPINDD67 pKa = 4.04GGGSPGGGTGGGDD80 pKa = 3.04GGYY83 pKa = 10.47AAARR87 pKa = 11.84SASRR91 pKa = 11.84SASQKK96 pKa = 10.54QNDD99 pKa = 3.41NTAALVDD106 pKa = 3.94QQRR109 pKa = 11.84KK110 pKa = 9.68LIDD113 pKa = 3.42AFGTQRR119 pKa = 11.84DD120 pKa = 4.03KK121 pKa = 11.56KK122 pKa = 10.66LGNINNSFTTSDD134 pKa = 3.43SLLMKK139 pKa = 10.59NYY141 pKa = 10.08GLAAEE146 pKa = 4.81GLAGTRR152 pKa = 11.84RR153 pKa = 11.84QNEE156 pKa = 4.08MAEE159 pKa = 4.22GDD161 pKa = 3.47ASFSNISNAVRR172 pKa = 11.84EE173 pKa = 4.21RR174 pKa = 11.84QNIADD179 pKa = 3.65QAASQGAGEE188 pKa = 4.21TDD190 pKa = 3.65LLRR193 pKa = 11.84SQLAAFRR200 pKa = 11.84NYY202 pKa = 10.24AANQGEE208 pKa = 4.65VNRR211 pKa = 11.84SFNDD215 pKa = 3.41TLQSINNSLTSLNSDD230 pKa = 3.18TSTSRR235 pKa = 11.84TNLFNQRR242 pKa = 11.84EE243 pKa = 4.35ADD245 pKa = 5.2RR246 pKa = 11.84EE247 pKa = 4.28AAWANYY253 pKa = 9.57ANQTSDD259 pKa = 2.93AWTQIANIEE268 pKa = 4.13NANTNTEE275 pKa = 3.51SDD277 pKa = 3.41MTVGYY282 pKa = 10.11NKK284 pKa = 10.46KK285 pKa = 8.79FTDD288 pKa = 3.62AGAQAAAAVKK298 pKa = 10.45NSYY301 pKa = 10.08SRR303 pKa = 11.84QAIPTGWNEE312 pKa = 3.54WGGKK316 pKa = 9.26QGTQEE321 pKa = 3.97RR322 pKa = 11.84ALTSSNRR329 pKa = 11.84AAAISLGGPVRR340 pKa = 11.84RR341 pKa = 11.84PEE343 pKa = 4.09GATLRR348 pKa = 11.84RR349 pKa = 11.84WEE351 pKa = 4.15GG352 pKa = 3.07
MM1 pKa = 7.65AKK3 pKa = 10.13KK4 pKa = 8.85KK5 pKa = 9.33TEE7 pKa = 4.38TIGEE11 pKa = 4.17RR12 pKa = 11.84NKK14 pKa = 10.79RR15 pKa = 11.84LGNAPGTGSVLKK27 pKa = 10.25PGYY30 pKa = 6.92TAPASPWGSLGNFLNPKK47 pKa = 8.77KK48 pKa = 10.03QVPVAPQNPRR58 pKa = 11.84TVTTAPINDD67 pKa = 4.04GGGSPGGGTGGGDD80 pKa = 3.04GGYY83 pKa = 10.47AAARR87 pKa = 11.84SASRR91 pKa = 11.84SASQKK96 pKa = 10.54QNDD99 pKa = 3.41NTAALVDD106 pKa = 3.94QQRR109 pKa = 11.84KK110 pKa = 9.68LIDD113 pKa = 3.42AFGTQRR119 pKa = 11.84DD120 pKa = 4.03KK121 pKa = 11.56KK122 pKa = 10.66LGNINNSFTTSDD134 pKa = 3.43SLLMKK139 pKa = 10.59NYY141 pKa = 10.08GLAAEE146 pKa = 4.81GLAGTRR152 pKa = 11.84RR153 pKa = 11.84QNEE156 pKa = 4.08MAEE159 pKa = 4.22GDD161 pKa = 3.47ASFSNISNAVRR172 pKa = 11.84EE173 pKa = 4.21RR174 pKa = 11.84QNIADD179 pKa = 3.65QAASQGAGEE188 pKa = 4.21TDD190 pKa = 3.65LLRR193 pKa = 11.84SQLAAFRR200 pKa = 11.84NYY202 pKa = 10.24AANQGEE208 pKa = 4.65VNRR211 pKa = 11.84SFNDD215 pKa = 3.41TLQSINNSLTSLNSDD230 pKa = 3.18TSTSRR235 pKa = 11.84TNLFNQRR242 pKa = 11.84EE243 pKa = 4.35ADD245 pKa = 5.2RR246 pKa = 11.84EE247 pKa = 4.28AAWANYY253 pKa = 9.57ANQTSDD259 pKa = 2.93AWTQIANIEE268 pKa = 4.13NANTNTEE275 pKa = 3.51SDD277 pKa = 3.41MTVGYY282 pKa = 10.11NKK284 pKa = 10.46KK285 pKa = 8.79FTDD288 pKa = 3.62AGAQAAAAVKK298 pKa = 10.45NSYY301 pKa = 10.08SRR303 pKa = 11.84QAIPTGWNEE312 pKa = 3.54WGGKK316 pKa = 9.26QGTQEE321 pKa = 3.97RR322 pKa = 11.84ALTSSNRR329 pKa = 11.84AAAISLGGPVRR340 pKa = 11.84RR341 pKa = 11.84PEE343 pKa = 4.09GATLRR348 pKa = 11.84RR349 pKa = 11.84WEE351 pKa = 4.15GG352 pKa = 3.07
Molecular weight: 37.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
17156 |
32 |
4491 |
350.1 |
38.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.154 ± 0.505 | 0.373 ± 0.12 |
6.499 ± 0.216 | 6.01 ± 0.342 |
3.241 ± 0.139 | 8.166 ± 0.359 |
1.399 ± 0.195 | 4.494 ± 0.249 |
4.162 ± 0.301 | 8.335 ± 0.274 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.536 ± 0.18 | 4.506 ± 0.236 |
5.077 ± 0.173 | 4.558 ± 0.212 |
5.59 ± 0.284 | 6.254 ± 0.239 |
6.96 ± 0.434 | 6.75 ± 0.24 |
1.9 ± 0.134 | 3.037 ± 0.166 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
