
Sweet potato leaf curl Canary virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 8.03
Get precalculated fractions of proteins
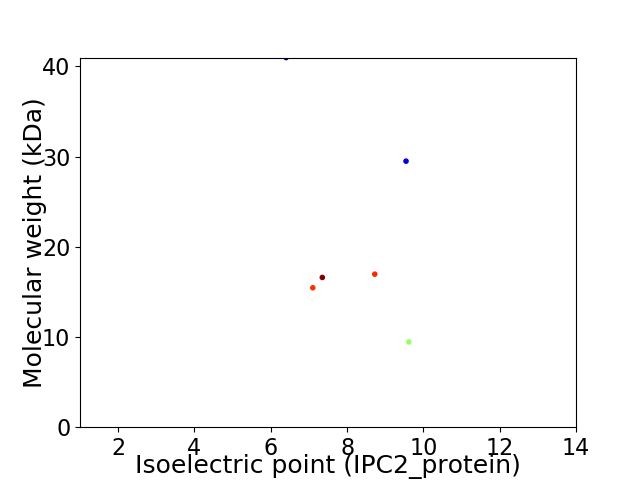
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
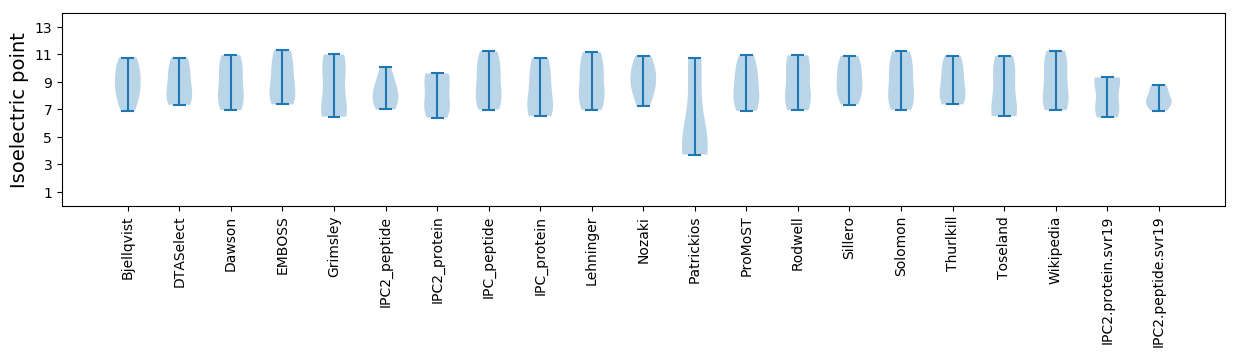
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C5H6W7|C5H6W7_9GEMI C4 OS=Sweet potato leaf curl Canary virus OX=652718 GN=C4 PE=3 SV=1
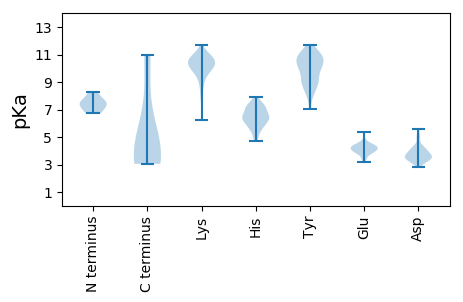
MM1 pKa = 7.53PRR3 pKa = 11.84KK4 pKa = 9.77QGFRR8 pKa = 11.84VQAKK12 pKa = 10.14NIFLTYY18 pKa = 9.16PKK20 pKa = 10.23CSLSKK25 pKa = 10.46EE26 pKa = 4.04QALEE30 pKa = 3.79QLRR33 pKa = 11.84ATHH36 pKa = 6.44CPSDD40 pKa = 3.38KK41 pKa = 11.01LFIRR45 pKa = 11.84VSQEE49 pKa = 3.19KK50 pKa = 9.64HH51 pKa = 4.73QDD53 pKa = 3.27GSLHH57 pKa = 5.85LHH59 pKa = 5.88VLIQFKK65 pKa = 10.82GKK67 pKa = 10.71AEE69 pKa = 4.33FKK71 pKa = 10.61NPRR74 pKa = 11.84HH75 pKa = 6.15FDD77 pKa = 2.89LHH79 pKa = 6.48HH80 pKa = 6.64PHH82 pKa = 6.75NSSQFHH88 pKa = 7.06PNFQAAKK95 pKa = 9.29SSSDD99 pKa = 3.13VKK101 pKa = 11.23SYY103 pKa = 10.8IEE105 pKa = 4.51KK106 pKa = 10.92DD107 pKa = 3.01GDD109 pKa = 3.73YY110 pKa = 10.84LDD112 pKa = 3.69WGEE115 pKa = 3.93FQIDD119 pKa = 3.21GRR121 pKa = 11.84SARR124 pKa = 11.84GGQQTANDD132 pKa = 3.91AAAEE136 pKa = 3.92ALNAGSKK143 pKa = 9.5EE144 pKa = 3.68AALQIIRR151 pKa = 11.84EE152 pKa = 4.21KK153 pKa = 11.03LPEE156 pKa = 4.12KK157 pKa = 11.02YY158 pKa = 9.91IFQYY162 pKa = 10.91HH163 pKa = 5.84NLVSNLDD170 pKa = 4.36RR171 pKa = 11.84IFSPPPAVYY180 pKa = 10.25CSPFSSSSFNNVPDD194 pKa = 5.62IISDD198 pKa = 3.84WAAEE202 pKa = 3.97NVMDD206 pKa = 5.1SAARR210 pKa = 11.84PDD212 pKa = 3.59RR213 pKa = 11.84PISIVIEE220 pKa = 4.52GPSRR224 pKa = 11.84IGKK227 pKa = 6.21TVWARR232 pKa = 11.84SLGPHH237 pKa = 6.71NYY239 pKa = 10.18LCGHH243 pKa = 7.38LDD245 pKa = 4.18LSPKK249 pKa = 10.09VYY251 pKa = 10.85SNSAWYY257 pKa = 10.34NVIDD261 pKa = 4.95DD262 pKa = 4.8VNPQYY267 pKa = 11.15LKK269 pKa = 10.65HH270 pKa = 6.26FKK272 pKa = 10.69EE273 pKa = 4.43FMGAQKK279 pKa = 10.56DD280 pKa = 3.62WQSNCKK286 pKa = 9.38YY287 pKa = 10.21GKK289 pKa = 9.18PVQIKK294 pKa = 10.41GGIPTIFLCNPGEE307 pKa = 4.28GSSFKK312 pKa = 10.83LWLDD316 pKa = 3.35KK317 pKa = 11.08PEE319 pKa = 4.24QEE321 pKa = 5.18ALKK324 pKa = 10.84NWALKK329 pKa = 10.36NAIFCDD335 pKa = 3.79VQSPFWVQEE344 pKa = 4.15EE345 pKa = 4.6VSGAGAITRR354 pKa = 11.84SSEE357 pKa = 3.86EE358 pKa = 4.0GQEE361 pKa = 4.12EE362 pKa = 4.77SSS364 pKa = 3.48
MM1 pKa = 7.53PRR3 pKa = 11.84KK4 pKa = 9.77QGFRR8 pKa = 11.84VQAKK12 pKa = 10.14NIFLTYY18 pKa = 9.16PKK20 pKa = 10.23CSLSKK25 pKa = 10.46EE26 pKa = 4.04QALEE30 pKa = 3.79QLRR33 pKa = 11.84ATHH36 pKa = 6.44CPSDD40 pKa = 3.38KK41 pKa = 11.01LFIRR45 pKa = 11.84VSQEE49 pKa = 3.19KK50 pKa = 9.64HH51 pKa = 4.73QDD53 pKa = 3.27GSLHH57 pKa = 5.85LHH59 pKa = 5.88VLIQFKK65 pKa = 10.82GKK67 pKa = 10.71AEE69 pKa = 4.33FKK71 pKa = 10.61NPRR74 pKa = 11.84HH75 pKa = 6.15FDD77 pKa = 2.89LHH79 pKa = 6.48HH80 pKa = 6.64PHH82 pKa = 6.75NSSQFHH88 pKa = 7.06PNFQAAKK95 pKa = 9.29SSSDD99 pKa = 3.13VKK101 pKa = 11.23SYY103 pKa = 10.8IEE105 pKa = 4.51KK106 pKa = 10.92DD107 pKa = 3.01GDD109 pKa = 3.73YY110 pKa = 10.84LDD112 pKa = 3.69WGEE115 pKa = 3.93FQIDD119 pKa = 3.21GRR121 pKa = 11.84SARR124 pKa = 11.84GGQQTANDD132 pKa = 3.91AAAEE136 pKa = 3.92ALNAGSKK143 pKa = 9.5EE144 pKa = 3.68AALQIIRR151 pKa = 11.84EE152 pKa = 4.21KK153 pKa = 11.03LPEE156 pKa = 4.12KK157 pKa = 11.02YY158 pKa = 9.91IFQYY162 pKa = 10.91HH163 pKa = 5.84NLVSNLDD170 pKa = 4.36RR171 pKa = 11.84IFSPPPAVYY180 pKa = 10.25CSPFSSSSFNNVPDD194 pKa = 5.62IISDD198 pKa = 3.84WAAEE202 pKa = 3.97NVMDD206 pKa = 5.1SAARR210 pKa = 11.84PDD212 pKa = 3.59RR213 pKa = 11.84PISIVIEE220 pKa = 4.52GPSRR224 pKa = 11.84IGKK227 pKa = 6.21TVWARR232 pKa = 11.84SLGPHH237 pKa = 6.71NYY239 pKa = 10.18LCGHH243 pKa = 7.38LDD245 pKa = 4.18LSPKK249 pKa = 10.09VYY251 pKa = 10.85SNSAWYY257 pKa = 10.34NVIDD261 pKa = 4.95DD262 pKa = 4.8VNPQYY267 pKa = 11.15LKK269 pKa = 10.65HH270 pKa = 6.26FKK272 pKa = 10.69EE273 pKa = 4.43FMGAQKK279 pKa = 10.56DD280 pKa = 3.62WQSNCKK286 pKa = 9.38YY287 pKa = 10.21GKK289 pKa = 9.18PVQIKK294 pKa = 10.41GGIPTIFLCNPGEE307 pKa = 4.28GSSFKK312 pKa = 10.83LWLDD316 pKa = 3.35KK317 pKa = 11.08PEE319 pKa = 4.24QEE321 pKa = 5.18ALKK324 pKa = 10.84NWALKK329 pKa = 10.36NAIFCDD335 pKa = 3.79VQSPFWVQEE344 pKa = 4.15EE345 pKa = 4.6VSGAGAITRR354 pKa = 11.84SSEE357 pKa = 3.86EE358 pKa = 4.0GQEE361 pKa = 4.12EE362 pKa = 4.77SSS364 pKa = 3.48
Molecular weight: 40.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C5HU40|C5HU40_9GEMI Protein V2 OS=Sweet potato leaf curl Canary virus OX=652718 GN=V2 PE=3 SV=1
MM1 pKa = 7.03GHH3 pKa = 7.23CISMFSSSSRR13 pKa = 11.84GKK15 pKa = 10.24QSSKK19 pKa = 8.25THH21 pKa = 7.04DD22 pKa = 2.81ISTYY26 pKa = 9.71IIHH29 pKa = 6.3ITPPNSIQISRR40 pKa = 11.84QLSPPRR46 pKa = 11.84MSSHH50 pKa = 5.23TSRR53 pKa = 11.84RR54 pKa = 11.84TEE56 pKa = 3.95TTSTGVNSRR65 pKa = 11.84STADD69 pKa = 3.5LLEE72 pKa = 4.45EE73 pKa = 4.23VSRR76 pKa = 11.84LLTTLPQRR84 pKa = 11.84HH85 pKa = 5.46
MM1 pKa = 7.03GHH3 pKa = 7.23CISMFSSSSRR13 pKa = 11.84GKK15 pKa = 10.24QSSKK19 pKa = 8.25THH21 pKa = 7.04DD22 pKa = 2.81ISTYY26 pKa = 9.71IIHH29 pKa = 6.3ITPPNSIQISRR40 pKa = 11.84QLSPPRR46 pKa = 11.84MSSHH50 pKa = 5.23TSRR53 pKa = 11.84RR54 pKa = 11.84TEE56 pKa = 3.95TTSTGVNSRR65 pKa = 11.84STADD69 pKa = 3.5LLEE72 pKa = 4.45EE73 pKa = 4.23VSRR76 pKa = 11.84LLTTLPQRR84 pKa = 11.84HH85 pKa = 5.46
Molecular weight: 9.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1127 |
85 |
364 |
187.8 |
21.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.324 ± 0.815 | 2.928 ± 0.775 |
4.969 ± 0.289 | 5.324 ± 0.704 |
4.17 ± 0.579 | 6.122 ± 0.398 |
3.549 ± 0.401 | 5.768 ± 0.534 |
6.034 ± 0.69 | 6.921 ± 0.934 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.218 ± 0.565 | 4.348 ± 0.554 |
6.122 ± 0.899 | 4.525 ± 0.647 |
7.453 ± 1.348 | 8.607 ± 1.399 |
5.235 ± 1.314 | 4.791 ± 0.95 |
1.775 ± 0.303 | 3.815 ± 0.619 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
