
Novipirellula artificiosorum
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Novipirellula
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
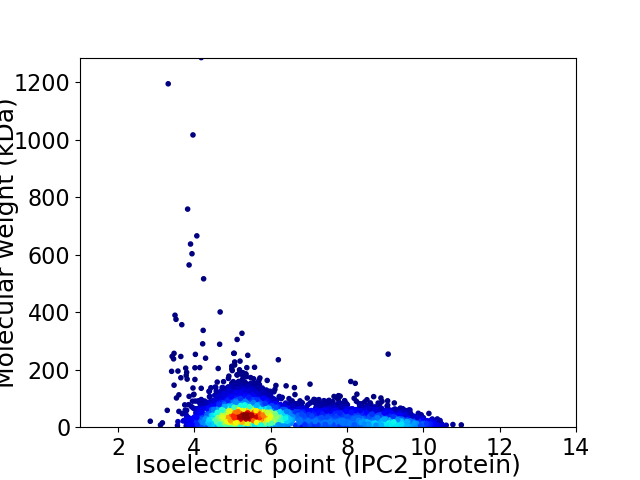
Virtual 2D-PAGE plot for 7087 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C6E2G2|A0A5C6E2G2_9BACT Uncharacterized protein OS=Novipirellula artificiosorum OX=2528016 GN=Poly41_12160 PE=4 SV=1
MM1 pKa = 7.42CLKK4 pKa = 10.47RR5 pKa = 11.84SRR7 pKa = 11.84SRR9 pKa = 11.84AWTTSEE15 pKa = 3.26WTATDD20 pKa = 4.11DD21 pKa = 4.98SATTDD26 pKa = 3.04EE27 pKa = 4.38DD28 pKa = 4.07TAVVMNVIANDD39 pKa = 3.73SDD41 pKa = 4.34PDD43 pKa = 4.18GDD45 pKa = 4.17SLLVDD50 pKa = 4.54SIGVAAHH57 pKa = 5.51GTIANNADD65 pKa = 3.17GTITYY70 pKa = 8.97TPDD73 pKa = 3.09ANYY76 pKa = 10.23HH77 pKa = 5.88GSDD80 pKa = 3.08SFTYY84 pKa = 10.16TLADD88 pKa = 3.54GNGGVDD94 pKa = 3.18TGTVMVSVTPVNDD107 pKa = 3.31APVAVDD113 pKa = 3.7DD114 pKa = 5.65DD115 pKa = 4.69YY116 pKa = 12.31VVDD119 pKa = 3.74QDD121 pKa = 3.55MSLVVPASGVLANDD135 pKa = 3.6SDD137 pKa = 3.98VDD139 pKa = 4.12ADD141 pKa = 4.0TLSAAIVGDD150 pKa = 4.08PSHH153 pKa = 6.94GDD155 pKa = 2.8VWLGEE160 pKa = 4.64DD161 pKa = 4.32GSFTYY166 pKa = 10.29TPEE169 pKa = 5.19DD170 pKa = 3.06GFYY173 pKa = 10.71GQDD176 pKa = 2.83SFTYY180 pKa = 9.79RR181 pKa = 11.84VQDD184 pKa = 3.33SSLVSDD190 pKa = 4.25TATVTIMVNRR200 pKa = 11.84VNSVPVANDD209 pKa = 3.02DD210 pKa = 4.3SYY212 pKa = 12.1DD213 pKa = 3.52VDD215 pKa = 3.85EE216 pKa = 5.16DD217 pKa = 3.8QPINVATPGVLGNDD231 pKa = 3.83RR232 pKa = 11.84DD233 pKa = 4.28DD234 pKa = 5.34DD235 pKa = 4.72GDD237 pKa = 5.1PITANIISGVSHH249 pKa = 6.45GQLNLNQDD257 pKa = 2.88GSFNYY262 pKa = 9.67TPEE265 pKa = 4.22ANFAGTDD272 pKa = 3.47SFSYY276 pKa = 9.98VANDD280 pKa = 3.45GFGNSDD286 pKa = 3.32QAIVTLTVNSVNDD299 pKa = 3.35VPTAEE304 pKa = 4.33SQSIVLPEE312 pKa = 5.11DD313 pKa = 3.37DD314 pKa = 4.79SKK316 pKa = 11.53PVTLQGSDD324 pKa = 3.73LDD326 pKa = 4.82GDD328 pKa = 4.08TLTYY332 pKa = 9.95TVTIGPTHH340 pKa = 6.06GTLAGTAPALTYY352 pKa = 10.5EE353 pKa = 4.15PAANYY358 pKa = 10.38SGLDD362 pKa = 3.18SFTFTVSDD370 pKa = 5.26GIAEE374 pKa = 4.2SAAATVSISVTSVNDD389 pKa = 3.66APILNQPIDD398 pKa = 4.21DD399 pKa = 4.73LDD401 pKa = 3.86VTEE404 pKa = 5.74DD405 pKa = 3.66SADD408 pKa = 3.62SVIDD412 pKa = 3.64LTGMFSDD419 pKa = 4.53IDD421 pKa = 3.84ASDD424 pKa = 4.08LVTLSVQANSNSSLVTASLLDD445 pKa = 3.61NVLTLDD451 pKa = 3.91YY452 pKa = 11.2QPDD455 pKa = 3.7QNGTATVTIRR465 pKa = 11.84GTDD468 pKa = 3.19GSGAYY473 pKa = 9.87AQDD476 pKa = 3.57SFLVTVTAVNDD487 pKa = 3.58APVAVDD493 pKa = 5.04DD494 pKa = 5.16LANTPMDD501 pKa = 3.9TPVVIDD507 pKa = 4.07VLDD510 pKa = 3.93NDD512 pKa = 4.11TDD514 pKa = 3.63VDD516 pKa = 4.27GEE518 pKa = 4.44SLSIAIIGSTNGTAVVNEE536 pKa = 4.1NGTVSFTPSASFSGLASFTYY556 pKa = 9.63TLNDD560 pKa = 3.61GEE562 pKa = 4.83LDD564 pKa = 3.98SNVATVTIDD573 pKa = 3.27VTPIASGPKK582 pKa = 8.56LAHH585 pKa = 5.74GVVTGVGSSGWTTVVLSHH603 pKa = 7.01TYY605 pKa = 11.51DD606 pKa = 3.39NMVVIATPNYY616 pKa = 9.94DD617 pKa = 3.28SSDD620 pKa = 3.6APGVTRR626 pKa = 11.84IQNASGNQFDD636 pKa = 4.36VRR638 pKa = 11.84VDD640 pKa = 3.59SAGGGSLSDD649 pKa = 3.4VRR651 pKa = 11.84VHH653 pKa = 5.44YY654 pKa = 10.53VVVEE658 pKa = 3.82AGFYY662 pKa = 10.64DD663 pKa = 3.7EE664 pKa = 5.66PGYY667 pKa = 11.33KK668 pKa = 8.83MEE670 pKa = 4.91AVTFNSTRR678 pKa = 11.84TDD680 pKa = 3.49EE681 pKa = 4.26NNSWVGEE688 pKa = 4.16SRR690 pKa = 11.84SYY692 pKa = 10.06LQSYY696 pKa = 7.18SQPVVLGQVMSYY708 pKa = 11.16NDD710 pKa = 4.34PDD712 pKa = 3.44WSVFWASGSSRR723 pKa = 11.84SAAPTSSALTVGKK736 pKa = 9.69HH737 pKa = 5.14VGEE740 pKa = 5.13DD741 pKa = 3.45SDD743 pKa = 3.79NTRR746 pKa = 11.84SDD748 pKa = 3.17EE749 pKa = 4.09TLGYY753 pKa = 9.95IVIEE757 pKa = 4.2TSTTGTAEE765 pKa = 4.07IEE767 pKa = 4.13GLRR770 pKa = 11.84YY771 pKa = 9.26VAALGGDD778 pKa = 4.41SIRR781 pKa = 11.84GVGDD785 pKa = 3.5SPAYY789 pKa = 9.91SYY791 pKa = 11.26SYY793 pKa = 10.83QAMPNSKK800 pKa = 8.06TAVASQAAMDD810 pKa = 4.42GGNGGWAVLYY820 pKa = 10.67GNDD823 pKa = 4.84PITPTGNTINFAIDD837 pKa = 3.04EE838 pKa = 4.41DD839 pKa = 3.94QARR842 pKa = 11.84DD843 pKa = 3.42TEE845 pKa = 4.71RR846 pKa = 11.84YY847 pKa = 7.4HH848 pKa = 5.07TTEE851 pKa = 3.42QVAYY855 pKa = 8.88FIIDD859 pKa = 3.57PPVEE863 pKa = 3.98EE864 pKa = 4.75LRR866 pKa = 11.84ASVTAMDD873 pKa = 3.87ASGDD877 pKa = 3.73GRR879 pKa = 11.84VTSMDD884 pKa = 3.26ALLVINHH891 pKa = 6.97LNTNRR896 pKa = 11.84SHH898 pKa = 7.64DD899 pKa = 4.16NPSHH903 pKa = 7.29LDD905 pKa = 3.57LSQDD909 pKa = 3.61GAVTPLDD916 pKa = 3.47ALVTINHH923 pKa = 6.67LNQRR927 pKa = 11.84TRR929 pKa = 11.84SAIQSRR935 pKa = 11.84SPITPQAVDD944 pKa = 3.18QWWAMDD950 pKa = 5.05DD951 pKa = 3.67EE952 pKa = 6.42DD953 pKa = 6.03DD954 pKa = 5.11NEE956 pKa = 4.74GSMDD960 pKa = 3.8EE961 pKa = 4.4ALLDD965 pKa = 3.66ILISS969 pKa = 3.64
MM1 pKa = 7.42CLKK4 pKa = 10.47RR5 pKa = 11.84SRR7 pKa = 11.84SRR9 pKa = 11.84AWTTSEE15 pKa = 3.26WTATDD20 pKa = 4.11DD21 pKa = 4.98SATTDD26 pKa = 3.04EE27 pKa = 4.38DD28 pKa = 4.07TAVVMNVIANDD39 pKa = 3.73SDD41 pKa = 4.34PDD43 pKa = 4.18GDD45 pKa = 4.17SLLVDD50 pKa = 4.54SIGVAAHH57 pKa = 5.51GTIANNADD65 pKa = 3.17GTITYY70 pKa = 8.97TPDD73 pKa = 3.09ANYY76 pKa = 10.23HH77 pKa = 5.88GSDD80 pKa = 3.08SFTYY84 pKa = 10.16TLADD88 pKa = 3.54GNGGVDD94 pKa = 3.18TGTVMVSVTPVNDD107 pKa = 3.31APVAVDD113 pKa = 3.7DD114 pKa = 5.65DD115 pKa = 4.69YY116 pKa = 12.31VVDD119 pKa = 3.74QDD121 pKa = 3.55MSLVVPASGVLANDD135 pKa = 3.6SDD137 pKa = 3.98VDD139 pKa = 4.12ADD141 pKa = 4.0TLSAAIVGDD150 pKa = 4.08PSHH153 pKa = 6.94GDD155 pKa = 2.8VWLGEE160 pKa = 4.64DD161 pKa = 4.32GSFTYY166 pKa = 10.29TPEE169 pKa = 5.19DD170 pKa = 3.06GFYY173 pKa = 10.71GQDD176 pKa = 2.83SFTYY180 pKa = 9.79RR181 pKa = 11.84VQDD184 pKa = 3.33SSLVSDD190 pKa = 4.25TATVTIMVNRR200 pKa = 11.84VNSVPVANDD209 pKa = 3.02DD210 pKa = 4.3SYY212 pKa = 12.1DD213 pKa = 3.52VDD215 pKa = 3.85EE216 pKa = 5.16DD217 pKa = 3.8QPINVATPGVLGNDD231 pKa = 3.83RR232 pKa = 11.84DD233 pKa = 4.28DD234 pKa = 5.34DD235 pKa = 4.72GDD237 pKa = 5.1PITANIISGVSHH249 pKa = 6.45GQLNLNQDD257 pKa = 2.88GSFNYY262 pKa = 9.67TPEE265 pKa = 4.22ANFAGTDD272 pKa = 3.47SFSYY276 pKa = 9.98VANDD280 pKa = 3.45GFGNSDD286 pKa = 3.32QAIVTLTVNSVNDD299 pKa = 3.35VPTAEE304 pKa = 4.33SQSIVLPEE312 pKa = 5.11DD313 pKa = 3.37DD314 pKa = 4.79SKK316 pKa = 11.53PVTLQGSDD324 pKa = 3.73LDD326 pKa = 4.82GDD328 pKa = 4.08TLTYY332 pKa = 9.95TVTIGPTHH340 pKa = 6.06GTLAGTAPALTYY352 pKa = 10.5EE353 pKa = 4.15PAANYY358 pKa = 10.38SGLDD362 pKa = 3.18SFTFTVSDD370 pKa = 5.26GIAEE374 pKa = 4.2SAAATVSISVTSVNDD389 pKa = 3.66APILNQPIDD398 pKa = 4.21DD399 pKa = 4.73LDD401 pKa = 3.86VTEE404 pKa = 5.74DD405 pKa = 3.66SADD408 pKa = 3.62SVIDD412 pKa = 3.64LTGMFSDD419 pKa = 4.53IDD421 pKa = 3.84ASDD424 pKa = 4.08LVTLSVQANSNSSLVTASLLDD445 pKa = 3.61NVLTLDD451 pKa = 3.91YY452 pKa = 11.2QPDD455 pKa = 3.7QNGTATVTIRR465 pKa = 11.84GTDD468 pKa = 3.19GSGAYY473 pKa = 9.87AQDD476 pKa = 3.57SFLVTVTAVNDD487 pKa = 3.58APVAVDD493 pKa = 5.04DD494 pKa = 5.16LANTPMDD501 pKa = 3.9TPVVIDD507 pKa = 4.07VLDD510 pKa = 3.93NDD512 pKa = 4.11TDD514 pKa = 3.63VDD516 pKa = 4.27GEE518 pKa = 4.44SLSIAIIGSTNGTAVVNEE536 pKa = 4.1NGTVSFTPSASFSGLASFTYY556 pKa = 9.63TLNDD560 pKa = 3.61GEE562 pKa = 4.83LDD564 pKa = 3.98SNVATVTIDD573 pKa = 3.27VTPIASGPKK582 pKa = 8.56LAHH585 pKa = 5.74GVVTGVGSSGWTTVVLSHH603 pKa = 7.01TYY605 pKa = 11.51DD606 pKa = 3.39NMVVIATPNYY616 pKa = 9.94DD617 pKa = 3.28SSDD620 pKa = 3.6APGVTRR626 pKa = 11.84IQNASGNQFDD636 pKa = 4.36VRR638 pKa = 11.84VDD640 pKa = 3.59SAGGGSLSDD649 pKa = 3.4VRR651 pKa = 11.84VHH653 pKa = 5.44YY654 pKa = 10.53VVVEE658 pKa = 3.82AGFYY662 pKa = 10.64DD663 pKa = 3.7EE664 pKa = 5.66PGYY667 pKa = 11.33KK668 pKa = 8.83MEE670 pKa = 4.91AVTFNSTRR678 pKa = 11.84TDD680 pKa = 3.49EE681 pKa = 4.26NNSWVGEE688 pKa = 4.16SRR690 pKa = 11.84SYY692 pKa = 10.06LQSYY696 pKa = 7.18SQPVVLGQVMSYY708 pKa = 11.16NDD710 pKa = 4.34PDD712 pKa = 3.44WSVFWASGSSRR723 pKa = 11.84SAAPTSSALTVGKK736 pKa = 9.69HH737 pKa = 5.14VGEE740 pKa = 5.13DD741 pKa = 3.45SDD743 pKa = 3.79NTRR746 pKa = 11.84SDD748 pKa = 3.17EE749 pKa = 4.09TLGYY753 pKa = 9.95IVIEE757 pKa = 4.2TSTTGTAEE765 pKa = 4.07IEE767 pKa = 4.13GLRR770 pKa = 11.84YY771 pKa = 9.26VAALGGDD778 pKa = 4.41SIRR781 pKa = 11.84GVGDD785 pKa = 3.5SPAYY789 pKa = 9.91SYY791 pKa = 11.26SYY793 pKa = 10.83QAMPNSKK800 pKa = 8.06TAVASQAAMDD810 pKa = 4.42GGNGGWAVLYY820 pKa = 10.67GNDD823 pKa = 4.84PITPTGNTINFAIDD837 pKa = 3.04EE838 pKa = 4.41DD839 pKa = 3.94QARR842 pKa = 11.84DD843 pKa = 3.42TEE845 pKa = 4.71RR846 pKa = 11.84YY847 pKa = 7.4HH848 pKa = 5.07TTEE851 pKa = 3.42QVAYY855 pKa = 8.88FIIDD859 pKa = 3.57PPVEE863 pKa = 3.98EE864 pKa = 4.75LRR866 pKa = 11.84ASVTAMDD873 pKa = 3.87ASGDD877 pKa = 3.73GRR879 pKa = 11.84VTSMDD884 pKa = 3.26ALLVINHH891 pKa = 6.97LNTNRR896 pKa = 11.84SHH898 pKa = 7.64DD899 pKa = 4.16NPSHH903 pKa = 7.29LDD905 pKa = 3.57LSQDD909 pKa = 3.61GAVTPLDD916 pKa = 3.47ALVTINHH923 pKa = 6.67LNQRR927 pKa = 11.84TRR929 pKa = 11.84SAIQSRR935 pKa = 11.84SPITPQAVDD944 pKa = 3.18QWWAMDD950 pKa = 5.05DD951 pKa = 3.67EE952 pKa = 6.42DD953 pKa = 6.03DD954 pKa = 5.11NEE956 pKa = 4.74GSMDD960 pKa = 3.8EE961 pKa = 4.4ALLDD965 pKa = 3.66ILISS969 pKa = 3.64
Molecular weight: 101.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C6D2F9|A0A5C6D2F9_9BACT OPT oligopeptide transporter protein OS=Novipirellula artificiosorum OX=2528016 GN=Poly41_64890 PE=4 SV=1
MM1 pKa = 7.38GFRR4 pKa = 11.84RR5 pKa = 11.84FRR7 pKa = 11.84LDD9 pKa = 2.87VVTRR13 pKa = 11.84LGVLVFSLSLSFLGTKK29 pKa = 9.4TSVVFVVLGMGFMQGMFSILSAVTWPRR56 pKa = 11.84FFGRR60 pKa = 11.84SHH62 pKa = 6.88LGAVSGMSTSIVVVGTAISRR82 pKa = 11.84RR83 pKa = 11.84SRR85 pKa = 3.2
MM1 pKa = 7.38GFRR4 pKa = 11.84RR5 pKa = 11.84FRR7 pKa = 11.84LDD9 pKa = 2.87VVTRR13 pKa = 11.84LGVLVFSLSLSFLGTKK29 pKa = 9.4TSVVFVVLGMGFMQGMFSILSAVTWPRR56 pKa = 11.84FFGRR60 pKa = 11.84SHH62 pKa = 6.88LGAVSGMSTSIVVVGTAISRR82 pKa = 11.84RR83 pKa = 11.84SRR85 pKa = 3.2
Molecular weight: 9.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2704480 |
29 |
11978 |
381.6 |
42.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.392 ± 0.031 | 1.185 ± 0.015 |
6.361 ± 0.037 | 6.015 ± 0.03 |
3.73 ± 0.018 | 7.67 ± 0.034 |
2.235 ± 0.018 | 5.084 ± 0.018 |
3.676 ± 0.032 | 9.439 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.334 ± 0.019 | 3.29 ± 0.026 |
5.171 ± 0.026 | 4.044 ± 0.022 |
6.592 ± 0.044 | 6.721 ± 0.025 |
5.718 ± 0.056 | 7.228 ± 0.027 |
1.583 ± 0.017 | 2.532 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
