
Hubei narna-like virus 17
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 9.26
Get precalculated fractions of proteins
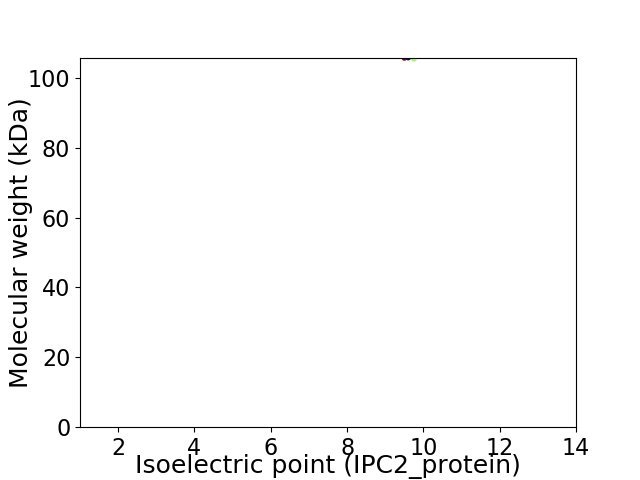
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIF7|A0A1L3KIF7_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 17 OX=1922947 PE=4 SV=1
MM1 pKa = 7.34VSPIGNLIVVRR12 pKa = 11.84PRR14 pKa = 11.84VYY16 pKa = 10.89TMGMTAAILRR26 pKa = 11.84RR27 pKa = 11.84MVHH30 pKa = 6.61RR31 pKa = 11.84NLAPGSLMLSLSRR44 pKa = 11.84SEE46 pKa = 3.68RR47 pKa = 11.84RR48 pKa = 11.84LRR50 pKa = 11.84QRR52 pKa = 11.84SEE54 pKa = 4.93VEE56 pKa = 4.14QQWLSDD62 pKa = 3.53FVRR65 pKa = 11.84FADD68 pKa = 3.88SAFALAIRR76 pKa = 11.84THH78 pKa = 6.31LRR80 pKa = 11.84QLQGDD85 pKa = 4.05DD86 pKa = 3.62VLAHH90 pKa = 6.23MKK92 pKa = 9.77TMARR96 pKa = 11.84LYY98 pKa = 11.1AFLAKK103 pKa = 10.77NEE105 pKa = 4.11VKK107 pKa = 10.56KK108 pKa = 10.62IASWWKK114 pKa = 8.76EE115 pKa = 3.47NCFQLLDD122 pKa = 3.83RR123 pKa = 11.84SLQAGDD129 pKa = 3.74EE130 pKa = 4.43ANNATGDD137 pKa = 3.91NEE139 pKa = 4.42VNTSFTRR146 pKa = 11.84SALAAASKK154 pKa = 10.75RR155 pKa = 11.84KK156 pKa = 8.94MKK158 pKa = 10.37RR159 pKa = 11.84ARR161 pKa = 11.84KK162 pKa = 9.29DD163 pKa = 3.14RR164 pKa = 11.84QRR166 pKa = 11.84VKK168 pKa = 10.95KK169 pKa = 10.91LMGTRR174 pKa = 11.84SSRR177 pKa = 11.84QVSSLFASLIRR188 pKa = 11.84SSGLQSFRR196 pKa = 11.84EE197 pKa = 5.07GIALGLSARR206 pKa = 11.84ILPAPILTEE215 pKa = 4.38PIVEE219 pKa = 4.17DD220 pKa = 3.37EE221 pKa = 3.92ARR223 pKa = 11.84KK224 pKa = 9.7LRR226 pKa = 11.84EE227 pKa = 3.95RR228 pKa = 11.84LTGRR232 pKa = 11.84PVVIPPEE239 pKa = 4.11DD240 pKa = 3.48AGIHH244 pKa = 4.41QSIGEE249 pKa = 4.18TMLRR253 pKa = 11.84ISNRR257 pKa = 11.84IKK259 pKa = 10.7RR260 pKa = 11.84FIKK263 pKa = 10.46VEE265 pKa = 3.86GRR267 pKa = 11.84KK268 pKa = 9.17IPLPTDD274 pKa = 3.38SKK276 pKa = 10.88IVSEE280 pKa = 4.36ADD282 pKa = 2.98THH284 pKa = 6.26EE285 pKa = 4.43VLTSRR290 pKa = 11.84CIAHH294 pKa = 6.91AAEE297 pKa = 4.52SSNGRR302 pKa = 11.84YY303 pKa = 6.96EE304 pKa = 4.05TFIGNVRR311 pKa = 11.84TVEE314 pKa = 3.85AKK316 pKa = 10.11PRR318 pKa = 11.84INKK321 pKa = 8.43TLFLAEE327 pKa = 4.24PGYY330 pKa = 10.01KK331 pKa = 9.86VRR333 pKa = 11.84VASIPPCSSMVTAGRR348 pKa = 11.84TVNQLLLRR356 pKa = 11.84AISKK360 pKa = 9.9AYY362 pKa = 8.69PAQTSLGIGTPKK374 pKa = 10.41EE375 pKa = 3.68FLAEE379 pKa = 4.18FKK381 pKa = 10.89KK382 pKa = 10.62CDD384 pKa = 3.52RR385 pKa = 11.84QCRR388 pKa = 11.84WLYY391 pKa = 10.16STDD394 pKa = 2.9LTAATDD400 pKa = 4.78HH401 pKa = 6.95IDD403 pKa = 2.98SSYY406 pKa = 11.83AMSAVEE412 pKa = 4.76DD413 pKa = 5.23LITEE417 pKa = 4.24MVNQGFTAAEE427 pKa = 4.22SVFWDD432 pKa = 4.12QLPEE436 pKa = 4.0MQTMAVCHH444 pKa = 5.31QNSRR448 pKa = 11.84EE449 pKa = 3.96PFMSFTMKK457 pKa = 10.44KK458 pKa = 9.25GVLMGKK464 pKa = 9.16PLTWPILTILQLTALEE480 pKa = 4.32HH481 pKa = 6.51AGLLARR487 pKa = 11.84SHH489 pKa = 6.51IVGDD493 pKa = 3.79DD494 pKa = 3.37ALVYY498 pKa = 10.63ASEE501 pKa = 4.46EE502 pKa = 4.2EE503 pKa = 4.1YY504 pKa = 11.42ADD506 pKa = 3.82YY507 pKa = 11.46VKK509 pKa = 9.82TAQAMGLVFNPSKK522 pKa = 8.02THH524 pKa = 5.14RR525 pKa = 11.84TRR527 pKa = 11.84SYY529 pKa = 11.25GILAQNVIKK538 pKa = 10.72LVGVPTTRR546 pKa = 11.84DD547 pKa = 3.28VKK549 pKa = 11.1DD550 pKa = 3.68KK551 pKa = 10.8IVRR554 pKa = 11.84TTEE557 pKa = 3.55NTLWAHH563 pKa = 6.11IEE565 pKa = 3.7EE566 pKa = 4.59AARR569 pKa = 11.84KK570 pKa = 8.92QGLVHH575 pKa = 7.1FGMDD579 pKa = 3.24NVRR582 pKa = 11.84SPRR585 pKa = 11.84RR586 pKa = 11.84YY587 pKa = 8.08EE588 pKa = 3.9SYY590 pKa = 11.12AFRR593 pKa = 11.84IEE595 pKa = 3.8QQIPYY600 pKa = 9.84VPEE603 pKa = 3.73LLAARR608 pKa = 11.84GQLRR612 pKa = 11.84KK613 pKa = 10.24DD614 pKa = 3.32PYY616 pKa = 11.64APFNLPSVRR625 pKa = 11.84RR626 pKa = 11.84LATLPRR632 pKa = 11.84WIRR635 pKa = 11.84DD636 pKa = 3.4RR637 pKa = 11.84VPNAFARR644 pKa = 11.84ANPQATKK651 pKa = 10.74VLLKK655 pKa = 10.45RR656 pKa = 11.84GVDD659 pKa = 3.01ISYY662 pKa = 9.48PVQFGGAAVMSVTTIKK678 pKa = 10.78RR679 pKa = 11.84MTKK682 pKa = 9.13RR683 pKa = 11.84HH684 pKa = 5.68LKK686 pKa = 9.57GAYY689 pKa = 9.63ALGRR693 pKa = 11.84YY694 pKa = 8.68ILANPTSYY702 pKa = 10.72TRR704 pKa = 11.84MSRR707 pKa = 11.84LMTTQATHH715 pKa = 6.01KK716 pKa = 9.31RR717 pKa = 11.84VTVTPVAITVEE728 pKa = 4.43KK729 pKa = 10.52VLEE732 pKa = 5.09DD733 pKa = 3.34IPTVPKK739 pKa = 10.94GKK741 pKa = 10.53GCDD744 pKa = 2.83IKK746 pKa = 8.62TTRR749 pKa = 11.84EE750 pKa = 3.8AVRR753 pKa = 11.84SITNDD758 pKa = 3.09ALVLLEE764 pKa = 4.17FAKK767 pKa = 10.74ASQVKK772 pKa = 10.35DD773 pKa = 3.49RR774 pKa = 11.84ITYY777 pKa = 9.58RR778 pKa = 11.84FEE780 pKa = 3.88KK781 pKa = 7.71TAHH784 pKa = 5.98MAACSHH790 pKa = 6.94PSMVLEE796 pKa = 4.15SFANKK801 pKa = 8.42ARR803 pKa = 11.84KK804 pKa = 8.52MKK806 pKa = 10.18VPRR809 pKa = 11.84PEE811 pKa = 3.64TVYY814 pKa = 11.15KK815 pKa = 10.24LWKK818 pKa = 9.75SLRR821 pKa = 11.84EE822 pKa = 3.94PNIDD826 pKa = 3.77YY827 pKa = 10.94DD828 pKa = 4.07PEE830 pKa = 4.12ALKK833 pKa = 10.8DD834 pKa = 3.34LWRR837 pKa = 11.84LSNATGATIVPTVSTNKK854 pKa = 9.82IEE856 pKa = 4.28TLRR859 pKa = 11.84LAKK862 pKa = 10.45YY863 pKa = 10.04SSLRR867 pKa = 11.84NTSAQNLDD875 pKa = 3.5TSTQDD880 pKa = 3.04TKK882 pKa = 10.94IIGNRR887 pKa = 11.84EE888 pKa = 3.84VSSSISLFGKK898 pKa = 9.94IATILAGNLEE908 pKa = 4.47LAQVQQITRR917 pKa = 11.84KK918 pKa = 8.17LTRR921 pKa = 11.84LQRR924 pKa = 11.84RR925 pKa = 11.84AVRR928 pKa = 11.84LGRR931 pKa = 11.84RR932 pKa = 11.84VRR934 pKa = 11.84ASAPTKK940 pKa = 10.72NSS942 pKa = 3.05
MM1 pKa = 7.34VSPIGNLIVVRR12 pKa = 11.84PRR14 pKa = 11.84VYY16 pKa = 10.89TMGMTAAILRR26 pKa = 11.84RR27 pKa = 11.84MVHH30 pKa = 6.61RR31 pKa = 11.84NLAPGSLMLSLSRR44 pKa = 11.84SEE46 pKa = 3.68RR47 pKa = 11.84RR48 pKa = 11.84LRR50 pKa = 11.84QRR52 pKa = 11.84SEE54 pKa = 4.93VEE56 pKa = 4.14QQWLSDD62 pKa = 3.53FVRR65 pKa = 11.84FADD68 pKa = 3.88SAFALAIRR76 pKa = 11.84THH78 pKa = 6.31LRR80 pKa = 11.84QLQGDD85 pKa = 4.05DD86 pKa = 3.62VLAHH90 pKa = 6.23MKK92 pKa = 9.77TMARR96 pKa = 11.84LYY98 pKa = 11.1AFLAKK103 pKa = 10.77NEE105 pKa = 4.11VKK107 pKa = 10.56KK108 pKa = 10.62IASWWKK114 pKa = 8.76EE115 pKa = 3.47NCFQLLDD122 pKa = 3.83RR123 pKa = 11.84SLQAGDD129 pKa = 3.74EE130 pKa = 4.43ANNATGDD137 pKa = 3.91NEE139 pKa = 4.42VNTSFTRR146 pKa = 11.84SALAAASKK154 pKa = 10.75RR155 pKa = 11.84KK156 pKa = 8.94MKK158 pKa = 10.37RR159 pKa = 11.84ARR161 pKa = 11.84KK162 pKa = 9.29DD163 pKa = 3.14RR164 pKa = 11.84QRR166 pKa = 11.84VKK168 pKa = 10.95KK169 pKa = 10.91LMGTRR174 pKa = 11.84SSRR177 pKa = 11.84QVSSLFASLIRR188 pKa = 11.84SSGLQSFRR196 pKa = 11.84EE197 pKa = 5.07GIALGLSARR206 pKa = 11.84ILPAPILTEE215 pKa = 4.38PIVEE219 pKa = 4.17DD220 pKa = 3.37EE221 pKa = 3.92ARR223 pKa = 11.84KK224 pKa = 9.7LRR226 pKa = 11.84EE227 pKa = 3.95RR228 pKa = 11.84LTGRR232 pKa = 11.84PVVIPPEE239 pKa = 4.11DD240 pKa = 3.48AGIHH244 pKa = 4.41QSIGEE249 pKa = 4.18TMLRR253 pKa = 11.84ISNRR257 pKa = 11.84IKK259 pKa = 10.7RR260 pKa = 11.84FIKK263 pKa = 10.46VEE265 pKa = 3.86GRR267 pKa = 11.84KK268 pKa = 9.17IPLPTDD274 pKa = 3.38SKK276 pKa = 10.88IVSEE280 pKa = 4.36ADD282 pKa = 2.98THH284 pKa = 6.26EE285 pKa = 4.43VLTSRR290 pKa = 11.84CIAHH294 pKa = 6.91AAEE297 pKa = 4.52SSNGRR302 pKa = 11.84YY303 pKa = 6.96EE304 pKa = 4.05TFIGNVRR311 pKa = 11.84TVEE314 pKa = 3.85AKK316 pKa = 10.11PRR318 pKa = 11.84INKK321 pKa = 8.43TLFLAEE327 pKa = 4.24PGYY330 pKa = 10.01KK331 pKa = 9.86VRR333 pKa = 11.84VASIPPCSSMVTAGRR348 pKa = 11.84TVNQLLLRR356 pKa = 11.84AISKK360 pKa = 9.9AYY362 pKa = 8.69PAQTSLGIGTPKK374 pKa = 10.41EE375 pKa = 3.68FLAEE379 pKa = 4.18FKK381 pKa = 10.89KK382 pKa = 10.62CDD384 pKa = 3.52RR385 pKa = 11.84QCRR388 pKa = 11.84WLYY391 pKa = 10.16STDD394 pKa = 2.9LTAATDD400 pKa = 4.78HH401 pKa = 6.95IDD403 pKa = 2.98SSYY406 pKa = 11.83AMSAVEE412 pKa = 4.76DD413 pKa = 5.23LITEE417 pKa = 4.24MVNQGFTAAEE427 pKa = 4.22SVFWDD432 pKa = 4.12QLPEE436 pKa = 4.0MQTMAVCHH444 pKa = 5.31QNSRR448 pKa = 11.84EE449 pKa = 3.96PFMSFTMKK457 pKa = 10.44KK458 pKa = 9.25GVLMGKK464 pKa = 9.16PLTWPILTILQLTALEE480 pKa = 4.32HH481 pKa = 6.51AGLLARR487 pKa = 11.84SHH489 pKa = 6.51IVGDD493 pKa = 3.79DD494 pKa = 3.37ALVYY498 pKa = 10.63ASEE501 pKa = 4.46EE502 pKa = 4.2EE503 pKa = 4.1YY504 pKa = 11.42ADD506 pKa = 3.82YY507 pKa = 11.46VKK509 pKa = 9.82TAQAMGLVFNPSKK522 pKa = 8.02THH524 pKa = 5.14RR525 pKa = 11.84TRR527 pKa = 11.84SYY529 pKa = 11.25GILAQNVIKK538 pKa = 10.72LVGVPTTRR546 pKa = 11.84DD547 pKa = 3.28VKK549 pKa = 11.1DD550 pKa = 3.68KK551 pKa = 10.8IVRR554 pKa = 11.84TTEE557 pKa = 3.55NTLWAHH563 pKa = 6.11IEE565 pKa = 3.7EE566 pKa = 4.59AARR569 pKa = 11.84KK570 pKa = 8.92QGLVHH575 pKa = 7.1FGMDD579 pKa = 3.24NVRR582 pKa = 11.84SPRR585 pKa = 11.84RR586 pKa = 11.84YY587 pKa = 8.08EE588 pKa = 3.9SYY590 pKa = 11.12AFRR593 pKa = 11.84IEE595 pKa = 3.8QQIPYY600 pKa = 9.84VPEE603 pKa = 3.73LLAARR608 pKa = 11.84GQLRR612 pKa = 11.84KK613 pKa = 10.24DD614 pKa = 3.32PYY616 pKa = 11.64APFNLPSVRR625 pKa = 11.84RR626 pKa = 11.84LATLPRR632 pKa = 11.84WIRR635 pKa = 11.84DD636 pKa = 3.4RR637 pKa = 11.84VPNAFARR644 pKa = 11.84ANPQATKK651 pKa = 10.74VLLKK655 pKa = 10.45RR656 pKa = 11.84GVDD659 pKa = 3.01ISYY662 pKa = 9.48PVQFGGAAVMSVTTIKK678 pKa = 10.78RR679 pKa = 11.84MTKK682 pKa = 9.13RR683 pKa = 11.84HH684 pKa = 5.68LKK686 pKa = 9.57GAYY689 pKa = 9.63ALGRR693 pKa = 11.84YY694 pKa = 8.68ILANPTSYY702 pKa = 10.72TRR704 pKa = 11.84MSRR707 pKa = 11.84LMTTQATHH715 pKa = 6.01KK716 pKa = 9.31RR717 pKa = 11.84VTVTPVAITVEE728 pKa = 4.43KK729 pKa = 10.52VLEE732 pKa = 5.09DD733 pKa = 3.34IPTVPKK739 pKa = 10.94GKK741 pKa = 10.53GCDD744 pKa = 2.83IKK746 pKa = 8.62TTRR749 pKa = 11.84EE750 pKa = 3.8AVRR753 pKa = 11.84SITNDD758 pKa = 3.09ALVLLEE764 pKa = 4.17FAKK767 pKa = 10.74ASQVKK772 pKa = 10.35DD773 pKa = 3.49RR774 pKa = 11.84ITYY777 pKa = 9.58RR778 pKa = 11.84FEE780 pKa = 3.88KK781 pKa = 7.71TAHH784 pKa = 5.98MAACSHH790 pKa = 6.94PSMVLEE796 pKa = 4.15SFANKK801 pKa = 8.42ARR803 pKa = 11.84KK804 pKa = 8.52MKK806 pKa = 10.18VPRR809 pKa = 11.84PEE811 pKa = 3.64TVYY814 pKa = 11.15KK815 pKa = 10.24LWKK818 pKa = 9.75SLRR821 pKa = 11.84EE822 pKa = 3.94PNIDD826 pKa = 3.77YY827 pKa = 10.94DD828 pKa = 4.07PEE830 pKa = 4.12ALKK833 pKa = 10.8DD834 pKa = 3.34LWRR837 pKa = 11.84LSNATGATIVPTVSTNKK854 pKa = 9.82IEE856 pKa = 4.28TLRR859 pKa = 11.84LAKK862 pKa = 10.45YY863 pKa = 10.04SSLRR867 pKa = 11.84NTSAQNLDD875 pKa = 3.5TSTQDD880 pKa = 3.04TKK882 pKa = 10.94IIGNRR887 pKa = 11.84EE888 pKa = 3.84VSSSISLFGKK898 pKa = 9.94IATILAGNLEE908 pKa = 4.47LAQVQQITRR917 pKa = 11.84KK918 pKa = 8.17LTRR921 pKa = 11.84LQRR924 pKa = 11.84RR925 pKa = 11.84AVRR928 pKa = 11.84LGRR931 pKa = 11.84RR932 pKa = 11.84VRR934 pKa = 11.84ASAPTKK940 pKa = 10.72NSS942 pKa = 3.05
Molecular weight: 105.66 kDa
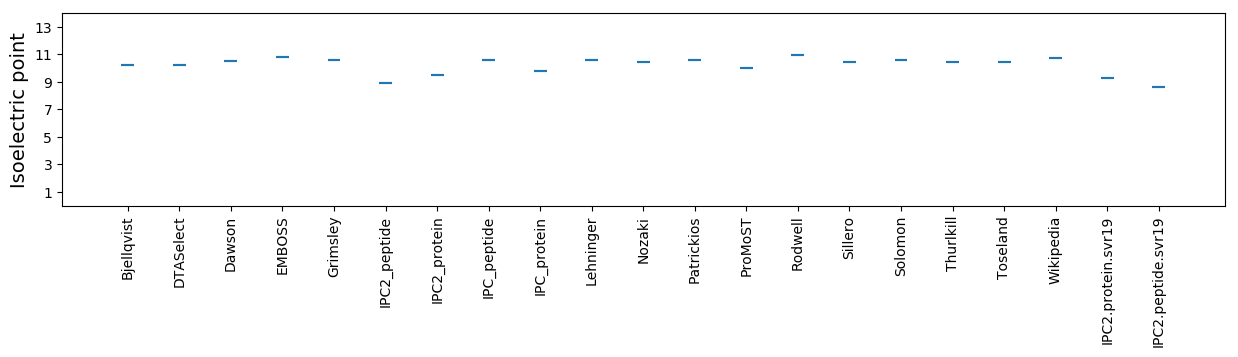
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIF7|A0A1L3KIF7_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 17 OX=1922947 PE=4 SV=1
MM1 pKa = 7.34VSPIGNLIVVRR12 pKa = 11.84PRR14 pKa = 11.84VYY16 pKa = 10.89TMGMTAAILRR26 pKa = 11.84RR27 pKa = 11.84MVHH30 pKa = 6.61RR31 pKa = 11.84NLAPGSLMLSLSRR44 pKa = 11.84SEE46 pKa = 3.68RR47 pKa = 11.84RR48 pKa = 11.84LRR50 pKa = 11.84QRR52 pKa = 11.84SEE54 pKa = 4.93VEE56 pKa = 4.14QQWLSDD62 pKa = 3.53FVRR65 pKa = 11.84FADD68 pKa = 3.88SAFALAIRR76 pKa = 11.84THH78 pKa = 6.31LRR80 pKa = 11.84QLQGDD85 pKa = 4.05DD86 pKa = 3.62VLAHH90 pKa = 6.23MKK92 pKa = 9.77TMARR96 pKa = 11.84LYY98 pKa = 11.1AFLAKK103 pKa = 10.77NEE105 pKa = 4.11VKK107 pKa = 10.56KK108 pKa = 10.62IASWWKK114 pKa = 8.76EE115 pKa = 3.47NCFQLLDD122 pKa = 3.83RR123 pKa = 11.84SLQAGDD129 pKa = 3.74EE130 pKa = 4.43ANNATGDD137 pKa = 3.91NEE139 pKa = 4.42VNTSFTRR146 pKa = 11.84SALAAASKK154 pKa = 10.75RR155 pKa = 11.84KK156 pKa = 8.94MKK158 pKa = 10.37RR159 pKa = 11.84ARR161 pKa = 11.84KK162 pKa = 9.29DD163 pKa = 3.14RR164 pKa = 11.84QRR166 pKa = 11.84VKK168 pKa = 10.95KK169 pKa = 10.91LMGTRR174 pKa = 11.84SSRR177 pKa = 11.84QVSSLFASLIRR188 pKa = 11.84SSGLQSFRR196 pKa = 11.84EE197 pKa = 5.07GIALGLSARR206 pKa = 11.84ILPAPILTEE215 pKa = 4.38PIVEE219 pKa = 4.17DD220 pKa = 3.37EE221 pKa = 3.92ARR223 pKa = 11.84KK224 pKa = 9.7LRR226 pKa = 11.84EE227 pKa = 3.95RR228 pKa = 11.84LTGRR232 pKa = 11.84PVVIPPEE239 pKa = 4.11DD240 pKa = 3.48AGIHH244 pKa = 4.41QSIGEE249 pKa = 4.18TMLRR253 pKa = 11.84ISNRR257 pKa = 11.84IKK259 pKa = 10.7RR260 pKa = 11.84FIKK263 pKa = 10.46VEE265 pKa = 3.86GRR267 pKa = 11.84KK268 pKa = 9.17IPLPTDD274 pKa = 3.38SKK276 pKa = 10.88IVSEE280 pKa = 4.36ADD282 pKa = 2.98THH284 pKa = 6.26EE285 pKa = 4.43VLTSRR290 pKa = 11.84CIAHH294 pKa = 6.91AAEE297 pKa = 4.52SSNGRR302 pKa = 11.84YY303 pKa = 6.96EE304 pKa = 4.05TFIGNVRR311 pKa = 11.84TVEE314 pKa = 3.85AKK316 pKa = 10.11PRR318 pKa = 11.84INKK321 pKa = 8.43TLFLAEE327 pKa = 4.24PGYY330 pKa = 10.01KK331 pKa = 9.86VRR333 pKa = 11.84VASIPPCSSMVTAGRR348 pKa = 11.84TVNQLLLRR356 pKa = 11.84AISKK360 pKa = 9.9AYY362 pKa = 8.69PAQTSLGIGTPKK374 pKa = 10.41EE375 pKa = 3.68FLAEE379 pKa = 4.18FKK381 pKa = 10.89KK382 pKa = 10.62CDD384 pKa = 3.52RR385 pKa = 11.84QCRR388 pKa = 11.84WLYY391 pKa = 10.16STDD394 pKa = 2.9LTAATDD400 pKa = 4.78HH401 pKa = 6.95IDD403 pKa = 2.98SSYY406 pKa = 11.83AMSAVEE412 pKa = 4.76DD413 pKa = 5.23LITEE417 pKa = 4.24MVNQGFTAAEE427 pKa = 4.22SVFWDD432 pKa = 4.12QLPEE436 pKa = 4.0MQTMAVCHH444 pKa = 5.31QNSRR448 pKa = 11.84EE449 pKa = 3.96PFMSFTMKK457 pKa = 10.44KK458 pKa = 9.25GVLMGKK464 pKa = 9.16PLTWPILTILQLTALEE480 pKa = 4.32HH481 pKa = 6.51AGLLARR487 pKa = 11.84SHH489 pKa = 6.51IVGDD493 pKa = 3.79DD494 pKa = 3.37ALVYY498 pKa = 10.63ASEE501 pKa = 4.46EE502 pKa = 4.2EE503 pKa = 4.1YY504 pKa = 11.42ADD506 pKa = 3.82YY507 pKa = 11.46VKK509 pKa = 9.82TAQAMGLVFNPSKK522 pKa = 8.02THH524 pKa = 5.14RR525 pKa = 11.84TRR527 pKa = 11.84SYY529 pKa = 11.25GILAQNVIKK538 pKa = 10.72LVGVPTTRR546 pKa = 11.84DD547 pKa = 3.28VKK549 pKa = 11.1DD550 pKa = 3.68KK551 pKa = 10.8IVRR554 pKa = 11.84TTEE557 pKa = 3.55NTLWAHH563 pKa = 6.11IEE565 pKa = 3.7EE566 pKa = 4.59AARR569 pKa = 11.84KK570 pKa = 8.92QGLVHH575 pKa = 7.1FGMDD579 pKa = 3.24NVRR582 pKa = 11.84SPRR585 pKa = 11.84RR586 pKa = 11.84YY587 pKa = 8.08EE588 pKa = 3.9SYY590 pKa = 11.12AFRR593 pKa = 11.84IEE595 pKa = 3.8QQIPYY600 pKa = 9.84VPEE603 pKa = 3.73LLAARR608 pKa = 11.84GQLRR612 pKa = 11.84KK613 pKa = 10.24DD614 pKa = 3.32PYY616 pKa = 11.64APFNLPSVRR625 pKa = 11.84RR626 pKa = 11.84LATLPRR632 pKa = 11.84WIRR635 pKa = 11.84DD636 pKa = 3.4RR637 pKa = 11.84VPNAFARR644 pKa = 11.84ANPQATKK651 pKa = 10.74VLLKK655 pKa = 10.45RR656 pKa = 11.84GVDD659 pKa = 3.01ISYY662 pKa = 9.48PVQFGGAAVMSVTTIKK678 pKa = 10.78RR679 pKa = 11.84MTKK682 pKa = 9.13RR683 pKa = 11.84HH684 pKa = 5.68LKK686 pKa = 9.57GAYY689 pKa = 9.63ALGRR693 pKa = 11.84YY694 pKa = 8.68ILANPTSYY702 pKa = 10.72TRR704 pKa = 11.84MSRR707 pKa = 11.84LMTTQATHH715 pKa = 6.01KK716 pKa = 9.31RR717 pKa = 11.84VTVTPVAITVEE728 pKa = 4.43KK729 pKa = 10.52VLEE732 pKa = 5.09DD733 pKa = 3.34IPTVPKK739 pKa = 10.94GKK741 pKa = 10.53GCDD744 pKa = 2.83IKK746 pKa = 8.62TTRR749 pKa = 11.84EE750 pKa = 3.8AVRR753 pKa = 11.84SITNDD758 pKa = 3.09ALVLLEE764 pKa = 4.17FAKK767 pKa = 10.74ASQVKK772 pKa = 10.35DD773 pKa = 3.49RR774 pKa = 11.84ITYY777 pKa = 9.58RR778 pKa = 11.84FEE780 pKa = 3.88KK781 pKa = 7.71TAHH784 pKa = 5.98MAACSHH790 pKa = 6.94PSMVLEE796 pKa = 4.15SFANKK801 pKa = 8.42ARR803 pKa = 11.84KK804 pKa = 8.52MKK806 pKa = 10.18VPRR809 pKa = 11.84PEE811 pKa = 3.64TVYY814 pKa = 11.15KK815 pKa = 10.24LWKK818 pKa = 9.75SLRR821 pKa = 11.84EE822 pKa = 3.94PNIDD826 pKa = 3.77YY827 pKa = 10.94DD828 pKa = 4.07PEE830 pKa = 4.12ALKK833 pKa = 10.8DD834 pKa = 3.34LWRR837 pKa = 11.84LSNATGATIVPTVSTNKK854 pKa = 9.82IEE856 pKa = 4.28TLRR859 pKa = 11.84LAKK862 pKa = 10.45YY863 pKa = 10.04SSLRR867 pKa = 11.84NTSAQNLDD875 pKa = 3.5TSTQDD880 pKa = 3.04TKK882 pKa = 10.94IIGNRR887 pKa = 11.84EE888 pKa = 3.84VSSSISLFGKK898 pKa = 9.94IATILAGNLEE908 pKa = 4.47LAQVQQITRR917 pKa = 11.84KK918 pKa = 8.17LTRR921 pKa = 11.84LQRR924 pKa = 11.84RR925 pKa = 11.84AVRR928 pKa = 11.84LGRR931 pKa = 11.84RR932 pKa = 11.84VRR934 pKa = 11.84ASAPTKK940 pKa = 10.72NSS942 pKa = 3.05
MM1 pKa = 7.34VSPIGNLIVVRR12 pKa = 11.84PRR14 pKa = 11.84VYY16 pKa = 10.89TMGMTAAILRR26 pKa = 11.84RR27 pKa = 11.84MVHH30 pKa = 6.61RR31 pKa = 11.84NLAPGSLMLSLSRR44 pKa = 11.84SEE46 pKa = 3.68RR47 pKa = 11.84RR48 pKa = 11.84LRR50 pKa = 11.84QRR52 pKa = 11.84SEE54 pKa = 4.93VEE56 pKa = 4.14QQWLSDD62 pKa = 3.53FVRR65 pKa = 11.84FADD68 pKa = 3.88SAFALAIRR76 pKa = 11.84THH78 pKa = 6.31LRR80 pKa = 11.84QLQGDD85 pKa = 4.05DD86 pKa = 3.62VLAHH90 pKa = 6.23MKK92 pKa = 9.77TMARR96 pKa = 11.84LYY98 pKa = 11.1AFLAKK103 pKa = 10.77NEE105 pKa = 4.11VKK107 pKa = 10.56KK108 pKa = 10.62IASWWKK114 pKa = 8.76EE115 pKa = 3.47NCFQLLDD122 pKa = 3.83RR123 pKa = 11.84SLQAGDD129 pKa = 3.74EE130 pKa = 4.43ANNATGDD137 pKa = 3.91NEE139 pKa = 4.42VNTSFTRR146 pKa = 11.84SALAAASKK154 pKa = 10.75RR155 pKa = 11.84KK156 pKa = 8.94MKK158 pKa = 10.37RR159 pKa = 11.84ARR161 pKa = 11.84KK162 pKa = 9.29DD163 pKa = 3.14RR164 pKa = 11.84QRR166 pKa = 11.84VKK168 pKa = 10.95KK169 pKa = 10.91LMGTRR174 pKa = 11.84SSRR177 pKa = 11.84QVSSLFASLIRR188 pKa = 11.84SSGLQSFRR196 pKa = 11.84EE197 pKa = 5.07GIALGLSARR206 pKa = 11.84ILPAPILTEE215 pKa = 4.38PIVEE219 pKa = 4.17DD220 pKa = 3.37EE221 pKa = 3.92ARR223 pKa = 11.84KK224 pKa = 9.7LRR226 pKa = 11.84EE227 pKa = 3.95RR228 pKa = 11.84LTGRR232 pKa = 11.84PVVIPPEE239 pKa = 4.11DD240 pKa = 3.48AGIHH244 pKa = 4.41QSIGEE249 pKa = 4.18TMLRR253 pKa = 11.84ISNRR257 pKa = 11.84IKK259 pKa = 10.7RR260 pKa = 11.84FIKK263 pKa = 10.46VEE265 pKa = 3.86GRR267 pKa = 11.84KK268 pKa = 9.17IPLPTDD274 pKa = 3.38SKK276 pKa = 10.88IVSEE280 pKa = 4.36ADD282 pKa = 2.98THH284 pKa = 6.26EE285 pKa = 4.43VLTSRR290 pKa = 11.84CIAHH294 pKa = 6.91AAEE297 pKa = 4.52SSNGRR302 pKa = 11.84YY303 pKa = 6.96EE304 pKa = 4.05TFIGNVRR311 pKa = 11.84TVEE314 pKa = 3.85AKK316 pKa = 10.11PRR318 pKa = 11.84INKK321 pKa = 8.43TLFLAEE327 pKa = 4.24PGYY330 pKa = 10.01KK331 pKa = 9.86VRR333 pKa = 11.84VASIPPCSSMVTAGRR348 pKa = 11.84TVNQLLLRR356 pKa = 11.84AISKK360 pKa = 9.9AYY362 pKa = 8.69PAQTSLGIGTPKK374 pKa = 10.41EE375 pKa = 3.68FLAEE379 pKa = 4.18FKK381 pKa = 10.89KK382 pKa = 10.62CDD384 pKa = 3.52RR385 pKa = 11.84QCRR388 pKa = 11.84WLYY391 pKa = 10.16STDD394 pKa = 2.9LTAATDD400 pKa = 4.78HH401 pKa = 6.95IDD403 pKa = 2.98SSYY406 pKa = 11.83AMSAVEE412 pKa = 4.76DD413 pKa = 5.23LITEE417 pKa = 4.24MVNQGFTAAEE427 pKa = 4.22SVFWDD432 pKa = 4.12QLPEE436 pKa = 4.0MQTMAVCHH444 pKa = 5.31QNSRR448 pKa = 11.84EE449 pKa = 3.96PFMSFTMKK457 pKa = 10.44KK458 pKa = 9.25GVLMGKK464 pKa = 9.16PLTWPILTILQLTALEE480 pKa = 4.32HH481 pKa = 6.51AGLLARR487 pKa = 11.84SHH489 pKa = 6.51IVGDD493 pKa = 3.79DD494 pKa = 3.37ALVYY498 pKa = 10.63ASEE501 pKa = 4.46EE502 pKa = 4.2EE503 pKa = 4.1YY504 pKa = 11.42ADD506 pKa = 3.82YY507 pKa = 11.46VKK509 pKa = 9.82TAQAMGLVFNPSKK522 pKa = 8.02THH524 pKa = 5.14RR525 pKa = 11.84TRR527 pKa = 11.84SYY529 pKa = 11.25GILAQNVIKK538 pKa = 10.72LVGVPTTRR546 pKa = 11.84DD547 pKa = 3.28VKK549 pKa = 11.1DD550 pKa = 3.68KK551 pKa = 10.8IVRR554 pKa = 11.84TTEE557 pKa = 3.55NTLWAHH563 pKa = 6.11IEE565 pKa = 3.7EE566 pKa = 4.59AARR569 pKa = 11.84KK570 pKa = 8.92QGLVHH575 pKa = 7.1FGMDD579 pKa = 3.24NVRR582 pKa = 11.84SPRR585 pKa = 11.84RR586 pKa = 11.84YY587 pKa = 8.08EE588 pKa = 3.9SYY590 pKa = 11.12AFRR593 pKa = 11.84IEE595 pKa = 3.8QQIPYY600 pKa = 9.84VPEE603 pKa = 3.73LLAARR608 pKa = 11.84GQLRR612 pKa = 11.84KK613 pKa = 10.24DD614 pKa = 3.32PYY616 pKa = 11.64APFNLPSVRR625 pKa = 11.84RR626 pKa = 11.84LATLPRR632 pKa = 11.84WIRR635 pKa = 11.84DD636 pKa = 3.4RR637 pKa = 11.84VPNAFARR644 pKa = 11.84ANPQATKK651 pKa = 10.74VLLKK655 pKa = 10.45RR656 pKa = 11.84GVDD659 pKa = 3.01ISYY662 pKa = 9.48PVQFGGAAVMSVTTIKK678 pKa = 10.78RR679 pKa = 11.84MTKK682 pKa = 9.13RR683 pKa = 11.84HH684 pKa = 5.68LKK686 pKa = 9.57GAYY689 pKa = 9.63ALGRR693 pKa = 11.84YY694 pKa = 8.68ILANPTSYY702 pKa = 10.72TRR704 pKa = 11.84MSRR707 pKa = 11.84LMTTQATHH715 pKa = 6.01KK716 pKa = 9.31RR717 pKa = 11.84VTVTPVAITVEE728 pKa = 4.43KK729 pKa = 10.52VLEE732 pKa = 5.09DD733 pKa = 3.34IPTVPKK739 pKa = 10.94GKK741 pKa = 10.53GCDD744 pKa = 2.83IKK746 pKa = 8.62TTRR749 pKa = 11.84EE750 pKa = 3.8AVRR753 pKa = 11.84SITNDD758 pKa = 3.09ALVLLEE764 pKa = 4.17FAKK767 pKa = 10.74ASQVKK772 pKa = 10.35DD773 pKa = 3.49RR774 pKa = 11.84ITYY777 pKa = 9.58RR778 pKa = 11.84FEE780 pKa = 3.88KK781 pKa = 7.71TAHH784 pKa = 5.98MAACSHH790 pKa = 6.94PSMVLEE796 pKa = 4.15SFANKK801 pKa = 8.42ARR803 pKa = 11.84KK804 pKa = 8.52MKK806 pKa = 10.18VPRR809 pKa = 11.84PEE811 pKa = 3.64TVYY814 pKa = 11.15KK815 pKa = 10.24LWKK818 pKa = 9.75SLRR821 pKa = 11.84EE822 pKa = 3.94PNIDD826 pKa = 3.77YY827 pKa = 10.94DD828 pKa = 4.07PEE830 pKa = 4.12ALKK833 pKa = 10.8DD834 pKa = 3.34LWRR837 pKa = 11.84LSNATGATIVPTVSTNKK854 pKa = 9.82IEE856 pKa = 4.28TLRR859 pKa = 11.84LAKK862 pKa = 10.45YY863 pKa = 10.04SSLRR867 pKa = 11.84NTSAQNLDD875 pKa = 3.5TSTQDD880 pKa = 3.04TKK882 pKa = 10.94IIGNRR887 pKa = 11.84EE888 pKa = 3.84VSSSISLFGKK898 pKa = 9.94IATILAGNLEE908 pKa = 4.47LAQVQQITRR917 pKa = 11.84KK918 pKa = 8.17LTRR921 pKa = 11.84LQRR924 pKa = 11.84RR925 pKa = 11.84AVRR928 pKa = 11.84LGRR931 pKa = 11.84RR932 pKa = 11.84VRR934 pKa = 11.84ASAPTKK940 pKa = 10.72NSS942 pKa = 3.05
Molecular weight: 105.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
942 |
942 |
942 |
942.0 |
105.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.873 ± 0.0 | 0.849 ± 0.0 |
3.822 ± 0.0 | 5.308 ± 0.0 |
2.866 ± 0.0 | 4.565 ± 0.0 |
1.805 ± 0.0 | 5.626 ± 0.0 |
6.157 ± 0.0 | 9.448 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.866 ± 0.0 | 3.503 ± 0.0 |
4.671 ± 0.0 | 3.715 ± 0.0 |
9.342 ± 0.0 | 7.431 ± 0.0 |
7.643 ± 0.0 | 7.006 ± 0.0 |
1.062 ± 0.0 | 2.442 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
