
Rhodotorula taiwanensis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Pucciniomycotina; Microbotryomycetes; Sporidiobolales; Sporidiobolaceae; Rhodotorula
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
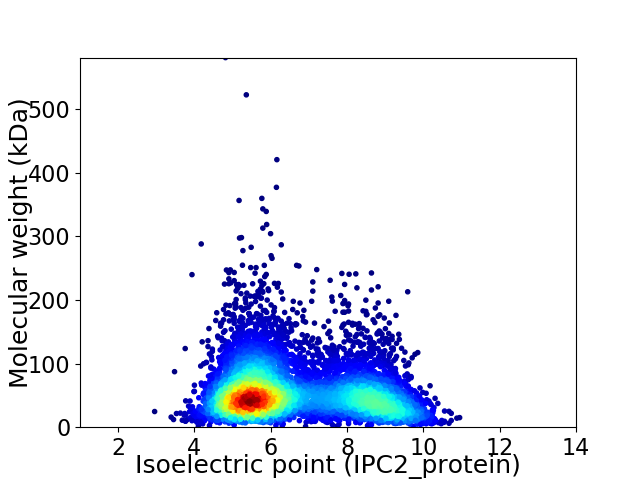
Virtual 2D-PAGE plot for 7012 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S5B4A1|A0A2S5B4A1_9BASI Uncharacterized protein OS=Rhodotorula taiwanensis OX=741276 GN=BMF94_5462 PE=4 SV=1
MM1 pKa = 7.4SVINLTDD8 pKa = 3.04VTVQNNPAGFTDD20 pKa = 5.21PYY22 pKa = 10.37QFQITFEE29 pKa = 4.44CVSPIQDD36 pKa = 3.53DD37 pKa = 4.78LEE39 pKa = 4.28WKK41 pKa = 8.51LTYY44 pKa = 10.43VGSAEE49 pKa = 4.01SDD51 pKa = 3.61SYY53 pKa = 11.69DD54 pKa = 4.17QEE56 pKa = 6.29LDD58 pKa = 3.15TCMVGPVPVGINSFPFEE75 pKa = 4.0AAAPLPSRR83 pKa = 11.84IPSSDD88 pKa = 3.79LIGVTVILLTCSYY101 pKa = 10.93NDD103 pKa = 3.35QEE105 pKa = 4.63FVRR108 pKa = 11.84IGYY111 pKa = 8.07YY112 pKa = 10.85VNTEE116 pKa = 3.93YY117 pKa = 11.4GDD119 pKa = 4.23PEE121 pKa = 5.04LKK123 pKa = 10.51AQYY126 pKa = 9.73DD127 pKa = 3.34ASLAEE132 pKa = 4.35DD133 pKa = 4.03AEE135 pKa = 4.59EE136 pKa = 4.56KK137 pKa = 10.64GIKK140 pKa = 10.38APDD143 pKa = 3.61PTQHH147 pKa = 6.42IDD149 pKa = 3.14KK150 pKa = 10.55LVRR153 pKa = 11.84NVLADD158 pKa = 3.5KK159 pKa = 10.88PRR161 pKa = 11.84VTKK164 pKa = 10.16FNIQWDD170 pKa = 4.45TPVAAVPTMSSAPIGGVTSEE190 pKa = 4.54ALAGAPAAAAPPPPFNPYY208 pKa = 9.6SAAAA212 pKa = 3.79
MM1 pKa = 7.4SVINLTDD8 pKa = 3.04VTVQNNPAGFTDD20 pKa = 5.21PYY22 pKa = 10.37QFQITFEE29 pKa = 4.44CVSPIQDD36 pKa = 3.53DD37 pKa = 4.78LEE39 pKa = 4.28WKK41 pKa = 8.51LTYY44 pKa = 10.43VGSAEE49 pKa = 4.01SDD51 pKa = 3.61SYY53 pKa = 11.69DD54 pKa = 4.17QEE56 pKa = 6.29LDD58 pKa = 3.15TCMVGPVPVGINSFPFEE75 pKa = 4.0AAAPLPSRR83 pKa = 11.84IPSSDD88 pKa = 3.79LIGVTVILLTCSYY101 pKa = 10.93NDD103 pKa = 3.35QEE105 pKa = 4.63FVRR108 pKa = 11.84IGYY111 pKa = 8.07YY112 pKa = 10.85VNTEE116 pKa = 3.93YY117 pKa = 11.4GDD119 pKa = 4.23PEE121 pKa = 5.04LKK123 pKa = 10.51AQYY126 pKa = 9.73DD127 pKa = 3.34ASLAEE132 pKa = 4.35DD133 pKa = 4.03AEE135 pKa = 4.59EE136 pKa = 4.56KK137 pKa = 10.64GIKK140 pKa = 10.38APDD143 pKa = 3.61PTQHH147 pKa = 6.42IDD149 pKa = 3.14KK150 pKa = 10.55LVRR153 pKa = 11.84NVLADD158 pKa = 3.5KK159 pKa = 10.88PRR161 pKa = 11.84VTKK164 pKa = 10.16FNIQWDD170 pKa = 4.45TPVAAVPTMSSAPIGGVTSEE190 pKa = 4.54ALAGAPAAAAPPPPFNPYY208 pKa = 9.6SAAAA212 pKa = 3.79
Molecular weight: 22.77 kDa
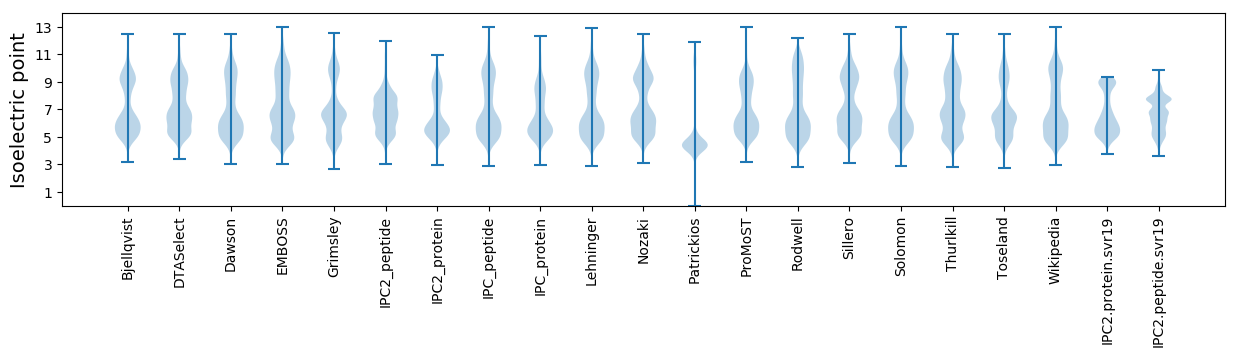
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S5BBQ0|A0A2S5BBQ0_9BASI Uncharacterized protein OS=Rhodotorula taiwanensis OX=741276 GN=BMF94_2752 PE=4 SV=1
MM1 pKa = 8.1DD2 pKa = 5.07KK3 pKa = 11.24GKK5 pKa = 11.15GKK7 pKa = 8.98ATEE10 pKa = 4.19LQPISLGVTLVADD23 pKa = 4.54GCPASEE29 pKa = 4.8GDD31 pKa = 5.1DD32 pKa = 3.75LAPSQLSHH40 pKa = 5.8VQAPATPPRR49 pKa = 11.84ASPLSVTSRR58 pKa = 11.84SSSFFDD64 pKa = 4.35DD65 pKa = 3.89EE66 pKa = 4.71PVPSTSSTPPQLSRR80 pKa = 11.84PAFVEE85 pKa = 4.18LVEE88 pKa = 4.43TCPTPPSAATTEE100 pKa = 3.98SSRR103 pKa = 11.84YY104 pKa = 8.65RR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84PLLQANPSLVSLALVNEE124 pKa = 4.49IGDD127 pKa = 4.58DD128 pKa = 3.76DD129 pKa = 4.07TPAAVNALDD138 pKa = 4.13EE139 pKa = 4.87RR140 pKa = 11.84PHH142 pKa = 6.21EE143 pKa = 5.33LPSAWQNDD151 pKa = 3.4THH153 pKa = 6.19SRR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84SRR159 pKa = 11.84QSRR162 pKa = 11.84RR163 pKa = 11.84STDD166 pKa = 3.14SARR169 pKa = 11.84TAAPLSEE176 pKa = 4.41LATAEE181 pKa = 4.11LVASILLQGPQVAAASTADD200 pKa = 3.63AQPHH204 pKa = 5.66LRR206 pKa = 11.84RR207 pKa = 11.84PSLDD211 pKa = 3.12TRR213 pKa = 11.84HH214 pKa = 6.42SSRR217 pKa = 11.84LSNDD221 pKa = 3.01PYY223 pKa = 11.04GVSSRR228 pKa = 11.84YY229 pKa = 9.18EE230 pKa = 4.02GSRR233 pKa = 11.84KK234 pKa = 9.85SPSLDD239 pKa = 3.05LRR241 pKa = 11.84QQSFSSSHH249 pKa = 5.24SLRR252 pKa = 11.84PLSTPQHH259 pKa = 6.28RR260 pKa = 11.84PSAPALLRR268 pKa = 11.84TRR270 pKa = 11.84PSTVTLVSWDD280 pKa = 4.15LSHH283 pKa = 7.26LPAPNTPRR291 pKa = 11.84EE292 pKa = 4.13RR293 pKa = 11.84PFKK296 pKa = 10.73VPFRR300 pKa = 11.84SVFLPTSYY308 pKa = 11.04LGGGKK313 pKa = 8.24GTGLYY318 pKa = 10.47GDD320 pKa = 3.55PCGPIEE326 pKa = 4.18RR327 pKa = 11.84KK328 pKa = 9.58VKK330 pKa = 10.42RR331 pKa = 11.84EE332 pKa = 3.42RR333 pKa = 11.84ARR335 pKa = 11.84PSNASGGSVGLGEE348 pKa = 4.26RR349 pKa = 11.84WRR351 pKa = 11.84RR352 pKa = 11.84WSGRR356 pKa = 11.84RR357 pKa = 11.84SSAMGYY363 pKa = 10.18GGRR366 pKa = 11.84TSWDD370 pKa = 3.6EE371 pKa = 3.97ASSIPPSATEE381 pKa = 4.32DD382 pKa = 3.46VGSLARR388 pKa = 11.84EE389 pKa = 4.23TEE391 pKa = 4.25RR392 pKa = 11.84PPSRR396 pKa = 11.84SSSLLRR402 pKa = 11.84RR403 pKa = 11.84FKK405 pKa = 10.64RR406 pKa = 11.84RR407 pKa = 11.84SWLSMPSSCEE417 pKa = 3.62VNGAPEE423 pKa = 4.01RR424 pKa = 11.84PNARR428 pKa = 11.84RR429 pKa = 11.84QSLTSALFGSRR440 pKa = 11.84RR441 pKa = 11.84SKK443 pKa = 10.71AASKK447 pKa = 10.6ASSEE451 pKa = 4.1GLEE454 pKa = 4.06TWVSVVVRR462 pKa = 4.49
MM1 pKa = 8.1DD2 pKa = 5.07KK3 pKa = 11.24GKK5 pKa = 11.15GKK7 pKa = 8.98ATEE10 pKa = 4.19LQPISLGVTLVADD23 pKa = 4.54GCPASEE29 pKa = 4.8GDD31 pKa = 5.1DD32 pKa = 3.75LAPSQLSHH40 pKa = 5.8VQAPATPPRR49 pKa = 11.84ASPLSVTSRR58 pKa = 11.84SSSFFDD64 pKa = 4.35DD65 pKa = 3.89EE66 pKa = 4.71PVPSTSSTPPQLSRR80 pKa = 11.84PAFVEE85 pKa = 4.18LVEE88 pKa = 4.43TCPTPPSAATTEE100 pKa = 3.98SSRR103 pKa = 11.84YY104 pKa = 8.65RR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84PLLQANPSLVSLALVNEE124 pKa = 4.49IGDD127 pKa = 4.58DD128 pKa = 3.76DD129 pKa = 4.07TPAAVNALDD138 pKa = 4.13EE139 pKa = 4.87RR140 pKa = 11.84PHH142 pKa = 6.21EE143 pKa = 5.33LPSAWQNDD151 pKa = 3.4THH153 pKa = 6.19SRR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84SRR159 pKa = 11.84QSRR162 pKa = 11.84RR163 pKa = 11.84STDD166 pKa = 3.14SARR169 pKa = 11.84TAAPLSEE176 pKa = 4.41LATAEE181 pKa = 4.11LVASILLQGPQVAAASTADD200 pKa = 3.63AQPHH204 pKa = 5.66LRR206 pKa = 11.84RR207 pKa = 11.84PSLDD211 pKa = 3.12TRR213 pKa = 11.84HH214 pKa = 6.42SSRR217 pKa = 11.84LSNDD221 pKa = 3.01PYY223 pKa = 11.04GVSSRR228 pKa = 11.84YY229 pKa = 9.18EE230 pKa = 4.02GSRR233 pKa = 11.84KK234 pKa = 9.85SPSLDD239 pKa = 3.05LRR241 pKa = 11.84QQSFSSSHH249 pKa = 5.24SLRR252 pKa = 11.84PLSTPQHH259 pKa = 6.28RR260 pKa = 11.84PSAPALLRR268 pKa = 11.84TRR270 pKa = 11.84PSTVTLVSWDD280 pKa = 4.15LSHH283 pKa = 7.26LPAPNTPRR291 pKa = 11.84EE292 pKa = 4.13RR293 pKa = 11.84PFKK296 pKa = 10.73VPFRR300 pKa = 11.84SVFLPTSYY308 pKa = 11.04LGGGKK313 pKa = 8.24GTGLYY318 pKa = 10.47GDD320 pKa = 3.55PCGPIEE326 pKa = 4.18RR327 pKa = 11.84KK328 pKa = 9.58VKK330 pKa = 10.42RR331 pKa = 11.84EE332 pKa = 3.42RR333 pKa = 11.84ARR335 pKa = 11.84PSNASGGSVGLGEE348 pKa = 4.26RR349 pKa = 11.84WRR351 pKa = 11.84RR352 pKa = 11.84WSGRR356 pKa = 11.84RR357 pKa = 11.84SSAMGYY363 pKa = 10.18GGRR366 pKa = 11.84TSWDD370 pKa = 3.6EE371 pKa = 3.97ASSIPPSATEE381 pKa = 4.32DD382 pKa = 3.46VGSLARR388 pKa = 11.84EE389 pKa = 4.23TEE391 pKa = 4.25RR392 pKa = 11.84PPSRR396 pKa = 11.84SSSLLRR402 pKa = 11.84RR403 pKa = 11.84FKK405 pKa = 10.64RR406 pKa = 11.84RR407 pKa = 11.84SWLSMPSSCEE417 pKa = 3.62VNGAPEE423 pKa = 4.01RR424 pKa = 11.84PNARR428 pKa = 11.84RR429 pKa = 11.84QSLTSALFGSRR440 pKa = 11.84RR441 pKa = 11.84SKK443 pKa = 10.71AASKK447 pKa = 10.6ASSEE451 pKa = 4.1GLEE454 pKa = 4.06TWVSVVVRR462 pKa = 4.49
Molecular weight: 49.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3853630 |
11 |
5360 |
549.6 |
59.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.484 ± 0.03 | 0.981 ± 0.009 |
5.79 ± 0.02 | 5.992 ± 0.023 |
3.232 ± 0.018 | 7.217 ± 0.025 |
2.222 ± 0.013 | 3.612 ± 0.019 |
4.029 ± 0.021 | 9.14 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.695 ± 0.009 | 2.578 ± 0.012 |
6.932 ± 0.034 | 3.768 ± 0.02 |
6.985 ± 0.023 | 8.834 ± 0.036 |
5.826 ± 0.016 | 6.154 ± 0.019 |
1.243 ± 0.01 | 2.287 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
