
Wenzhou sobemo-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.69
Get precalculated fractions of proteins
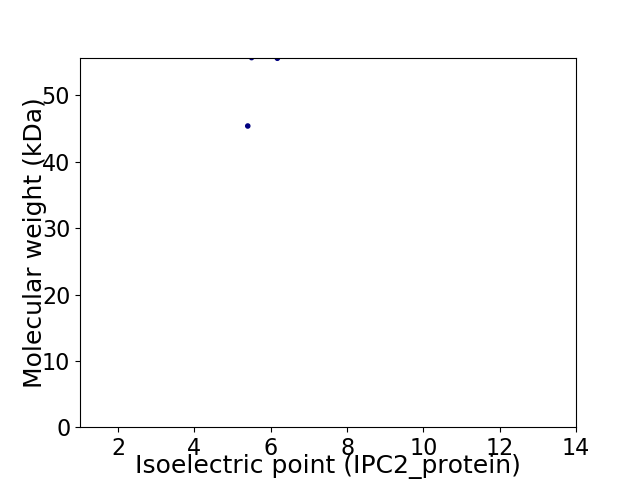
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEF3|A0A1L3KEF3_9VIRU Uncharacterized protein OS=Wenzhou sobemo-like virus 2 OX=1923658 PE=4 SV=1
MM1 pKa = 7.43RR2 pKa = 11.84TRR4 pKa = 11.84IKK6 pKa = 10.39IKK8 pKa = 10.33SDD10 pKa = 2.81VNTFVRR16 pKa = 11.84EE17 pKa = 3.85EE18 pKa = 4.35FIRR21 pKa = 11.84ALQDD25 pKa = 2.92IDD27 pKa = 4.37PNSSPGCCEE36 pKa = 3.48MKK38 pKa = 10.76VYY40 pKa = 8.97GTTNRR45 pKa = 11.84EE46 pKa = 3.87CLGIQGGPEE55 pKa = 4.13GRR57 pKa = 11.84YY58 pKa = 7.56MDD60 pKa = 4.2PVRR63 pKa = 11.84VEE65 pKa = 3.37ALYY68 pKa = 11.1RR69 pKa = 11.84MVIGRR74 pKa = 11.84IQNLFEE80 pKa = 4.45GNYY83 pKa = 10.18VSDD86 pKa = 3.84NLLTFVKK93 pKa = 10.57DD94 pKa = 3.67EE95 pKa = 4.02PTKK98 pKa = 10.57LSKK101 pKa = 10.52IEE103 pKa = 3.96TKK105 pKa = 9.68RR106 pKa = 11.84WRR108 pKa = 11.84LISGVSLVDD117 pKa = 3.27TMVDD121 pKa = 3.83RR122 pKa = 11.84ILLQFLFEE130 pKa = 4.21QYY132 pKa = 9.38MDD134 pKa = 3.84RR135 pKa = 11.84LGTTPVAIGWNPYY148 pKa = 7.69TSAYY152 pKa = 9.92LFSIVMTPSEE162 pKa = 4.34PKK164 pKa = 9.78KK165 pKa = 10.43VHH167 pKa = 5.51PQYY170 pKa = 11.59VAMDD174 pKa = 4.04KK175 pKa = 11.04SCWDD179 pKa = 3.18WTVQTWLLDD188 pKa = 4.03AIMQVFIGAVVEE200 pKa = 4.43PPDD203 pKa = 2.82WWVYY207 pKa = 9.42LLKK210 pKa = 10.22TRR212 pKa = 11.84FYY214 pKa = 11.12CLFEE218 pKa = 5.44APTFEE223 pKa = 4.28FQDD226 pKa = 3.49GFTIKK231 pKa = 10.32QKK233 pKa = 10.81EE234 pKa = 4.03PGIMKK239 pKa = 9.8SGCYY243 pKa = 6.2MTLMANSIAQVLLDD257 pKa = 4.18SLCAQAAKK265 pKa = 10.1IDD267 pKa = 3.39VHH269 pKa = 7.28YY270 pKa = 9.98PFWCCGDD277 pKa = 3.88DD278 pKa = 4.12TIQMLPSDD286 pKa = 4.49FRR288 pKa = 11.84QLVDD292 pKa = 3.91YY293 pKa = 10.25INAMEE298 pKa = 4.58RR299 pKa = 11.84YY300 pKa = 8.93SYY302 pKa = 10.37NLKK305 pKa = 10.42VSYY308 pKa = 10.21HH309 pKa = 7.02DD310 pKa = 3.24EE311 pKa = 4.47AEE313 pKa = 4.3FVGSIMTRR321 pKa = 11.84KK322 pKa = 10.13GPVPAYY328 pKa = 8.74QAKK331 pKa = 9.93HH332 pKa = 5.3CWLLQRR338 pKa = 11.84LTLKK342 pKa = 10.82PEE344 pKa = 4.02DD345 pKa = 3.62QIATLRR351 pKa = 11.84SYY353 pKa = 11.25QIMYY357 pKa = 10.55ANEE360 pKa = 4.07PEE362 pKa = 4.31VLAKK366 pKa = 10.0IRR368 pKa = 11.84GLIDD372 pKa = 2.76KK373 pKa = 9.86WNIPEE378 pKa = 4.26AYY380 pKa = 7.94ITAEE384 pKa = 4.25EE385 pKa = 4.17LRR387 pKa = 11.84EE388 pKa = 4.02LQQSS392 pKa = 3.39
MM1 pKa = 7.43RR2 pKa = 11.84TRR4 pKa = 11.84IKK6 pKa = 10.39IKK8 pKa = 10.33SDD10 pKa = 2.81VNTFVRR16 pKa = 11.84EE17 pKa = 3.85EE18 pKa = 4.35FIRR21 pKa = 11.84ALQDD25 pKa = 2.92IDD27 pKa = 4.37PNSSPGCCEE36 pKa = 3.48MKK38 pKa = 10.76VYY40 pKa = 8.97GTTNRR45 pKa = 11.84EE46 pKa = 3.87CLGIQGGPEE55 pKa = 4.13GRR57 pKa = 11.84YY58 pKa = 7.56MDD60 pKa = 4.2PVRR63 pKa = 11.84VEE65 pKa = 3.37ALYY68 pKa = 11.1RR69 pKa = 11.84MVIGRR74 pKa = 11.84IQNLFEE80 pKa = 4.45GNYY83 pKa = 10.18VSDD86 pKa = 3.84NLLTFVKK93 pKa = 10.57DD94 pKa = 3.67EE95 pKa = 4.02PTKK98 pKa = 10.57LSKK101 pKa = 10.52IEE103 pKa = 3.96TKK105 pKa = 9.68RR106 pKa = 11.84WRR108 pKa = 11.84LISGVSLVDD117 pKa = 3.27TMVDD121 pKa = 3.83RR122 pKa = 11.84ILLQFLFEE130 pKa = 4.21QYY132 pKa = 9.38MDD134 pKa = 3.84RR135 pKa = 11.84LGTTPVAIGWNPYY148 pKa = 7.69TSAYY152 pKa = 9.92LFSIVMTPSEE162 pKa = 4.34PKK164 pKa = 9.78KK165 pKa = 10.43VHH167 pKa = 5.51PQYY170 pKa = 11.59VAMDD174 pKa = 4.04KK175 pKa = 11.04SCWDD179 pKa = 3.18WTVQTWLLDD188 pKa = 4.03AIMQVFIGAVVEE200 pKa = 4.43PPDD203 pKa = 2.82WWVYY207 pKa = 9.42LLKK210 pKa = 10.22TRR212 pKa = 11.84FYY214 pKa = 11.12CLFEE218 pKa = 5.44APTFEE223 pKa = 4.28FQDD226 pKa = 3.49GFTIKK231 pKa = 10.32QKK233 pKa = 10.81EE234 pKa = 4.03PGIMKK239 pKa = 9.8SGCYY243 pKa = 6.2MTLMANSIAQVLLDD257 pKa = 4.18SLCAQAAKK265 pKa = 10.1IDD267 pKa = 3.39VHH269 pKa = 7.28YY270 pKa = 9.98PFWCCGDD277 pKa = 3.88DD278 pKa = 4.12TIQMLPSDD286 pKa = 4.49FRR288 pKa = 11.84QLVDD292 pKa = 3.91YY293 pKa = 10.25INAMEE298 pKa = 4.58RR299 pKa = 11.84YY300 pKa = 8.93SYY302 pKa = 10.37NLKK305 pKa = 10.42VSYY308 pKa = 10.21HH309 pKa = 7.02DD310 pKa = 3.24EE311 pKa = 4.47AEE313 pKa = 4.3FVGSIMTRR321 pKa = 11.84KK322 pKa = 10.13GPVPAYY328 pKa = 8.74QAKK331 pKa = 9.93HH332 pKa = 5.3CWLLQRR338 pKa = 11.84LTLKK342 pKa = 10.82PEE344 pKa = 4.02DD345 pKa = 3.62QIATLRR351 pKa = 11.84SYY353 pKa = 11.25QIMYY357 pKa = 10.55ANEE360 pKa = 4.07PEE362 pKa = 4.31VLAKK366 pKa = 10.0IRR368 pKa = 11.84GLIDD372 pKa = 2.76KK373 pKa = 9.86WNIPEE378 pKa = 4.26AYY380 pKa = 7.94ITAEE384 pKa = 4.25EE385 pKa = 4.17LRR387 pKa = 11.84EE388 pKa = 4.02LQQSS392 pKa = 3.39
Molecular weight: 45.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEF3|A0A1L3KEF3_9VIRU Uncharacterized protein OS=Wenzhou sobemo-like virus 2 OX=1923658 PE=4 SV=1
MM1 pKa = 7.0STVAPDD7 pKa = 3.55GTFMRR12 pKa = 11.84NEE14 pKa = 3.99AYY16 pKa = 10.75VPGSEE21 pKa = 4.22YY22 pKa = 10.56QRR24 pKa = 11.84TVDD27 pKa = 5.31GIPKK31 pKa = 8.31FQCQITDD38 pKa = 3.21KK39 pKa = 10.76FGSSVLGYY47 pKa = 8.43ATWVGDD53 pKa = 3.94YY54 pKa = 10.39IVTPTHH60 pKa = 6.11VVATANTDD68 pKa = 3.17HH69 pKa = 6.89VLLLTRR75 pKa = 11.84NGAVSVPTDD84 pKa = 3.04QWRR87 pKa = 11.84HH88 pKa = 4.93LEE90 pKa = 4.13YY91 pKa = 11.03ADD93 pKa = 4.24VSYY96 pKa = 11.1LPYY99 pKa = 10.19EE100 pKa = 3.9QRR102 pKa = 11.84TYY104 pKa = 11.68SPLQLSKK111 pKa = 10.62GTVYY115 pKa = 10.8PPGKK119 pKa = 9.57EE120 pKa = 3.99VNNLFVTATTSTVFTTGIIRR140 pKa = 11.84EE141 pKa = 4.38HH142 pKa = 6.47PTDD145 pKa = 3.61ALYY148 pKa = 9.05VTYY151 pKa = 10.31EE152 pKa = 4.15GSTKK156 pKa = 10.25PGFSGVPYY164 pKa = 10.31VSNKK168 pKa = 9.52KK169 pKa = 9.74AVAMHH174 pKa = 6.89LGSNSYY180 pKa = 10.99GKK182 pKa = 10.25GIKK185 pKa = 9.98YY186 pKa = 9.97SWILIMIYY194 pKa = 9.68EE195 pKa = 4.5AEE197 pKa = 4.18NPNQNVRR204 pKa = 11.84HH205 pKa = 5.64YY206 pKa = 11.0SDD208 pKa = 3.18EE209 pKa = 4.24TEE211 pKa = 3.81KK212 pKa = 11.16VFIRR216 pKa = 11.84EE217 pKa = 4.22STGSDD222 pKa = 2.97TYY224 pKa = 11.52EE225 pKa = 4.03KK226 pKa = 10.72LIAQALEE233 pKa = 3.96RR234 pKa = 11.84MSQGEE239 pKa = 4.5RR240 pKa = 11.84YY241 pKa = 9.16FKK243 pKa = 10.18RR244 pKa = 11.84AGYY247 pKa = 10.23RR248 pKa = 11.84GMDD251 pKa = 2.85SYY253 pKa = 11.84LFNNSDD259 pKa = 3.18GTMMEE264 pKa = 4.28ITPHH268 pKa = 5.32TRR270 pKa = 11.84QHH272 pKa = 4.67MMAFNEE278 pKa = 4.01YY279 pKa = 10.92DD280 pKa = 3.27NDD282 pKa = 4.04AEE284 pKa = 4.31MMSGGYY290 pKa = 8.66RR291 pKa = 11.84RR292 pKa = 11.84KK293 pKa = 9.88RR294 pKa = 11.84SRR296 pKa = 11.84KK297 pKa = 9.12APSGWGEE304 pKa = 4.14LEE306 pKa = 3.6EE307 pKa = 4.21LAPVRR312 pKa = 11.84KK313 pKa = 10.06GKK315 pKa = 10.92GKK317 pKa = 10.39GGKK320 pKa = 8.6FRR322 pKa = 11.84YY323 pKa = 9.69DD324 pKa = 3.24EE325 pKa = 4.5PPAFKK330 pKa = 10.58GKK332 pKa = 9.89SWVDD336 pKa = 3.13MMEE339 pKa = 3.92EE340 pKa = 3.54SDD342 pKa = 4.24YY343 pKa = 11.68EE344 pKa = 4.17EE345 pKa = 4.94EE346 pKa = 4.72NADD349 pKa = 5.67DD350 pKa = 4.45SNQDD354 pKa = 3.02QPMVSQVKK362 pKa = 9.63NKK364 pKa = 10.21KK365 pKa = 9.7AIKK368 pKa = 10.21KK369 pKa = 10.06LGLDD373 pKa = 3.77NVPQTAMLVPDD384 pKa = 4.11ISRR387 pKa = 11.84PTVPVYY393 pKa = 10.75SDD395 pKa = 3.41YY396 pKa = 11.54PEE398 pKa = 4.03VVKK401 pKa = 10.09TSKK404 pKa = 10.83NVVSPPDD411 pKa = 3.25VEE413 pKa = 5.55APGKK417 pKa = 7.45TQTAEE422 pKa = 3.89EE423 pKa = 4.31VVIPLRR429 pKa = 11.84TTTGSLPTPNFNLDD443 pKa = 3.57HH444 pKa = 7.06LSNTAAPPLTLAQLGEE460 pKa = 4.38ALPLILEE467 pKa = 4.46SLNSLSNRR475 pKa = 11.84LQNPRR480 pKa = 11.84RR481 pKa = 11.84RR482 pKa = 11.84RR483 pKa = 11.84SRR485 pKa = 11.84KK486 pKa = 9.17PSAKK490 pKa = 9.65PSNSCAQEE498 pKa = 3.64
MM1 pKa = 7.0STVAPDD7 pKa = 3.55GTFMRR12 pKa = 11.84NEE14 pKa = 3.99AYY16 pKa = 10.75VPGSEE21 pKa = 4.22YY22 pKa = 10.56QRR24 pKa = 11.84TVDD27 pKa = 5.31GIPKK31 pKa = 8.31FQCQITDD38 pKa = 3.21KK39 pKa = 10.76FGSSVLGYY47 pKa = 8.43ATWVGDD53 pKa = 3.94YY54 pKa = 10.39IVTPTHH60 pKa = 6.11VVATANTDD68 pKa = 3.17HH69 pKa = 6.89VLLLTRR75 pKa = 11.84NGAVSVPTDD84 pKa = 3.04QWRR87 pKa = 11.84HH88 pKa = 4.93LEE90 pKa = 4.13YY91 pKa = 11.03ADD93 pKa = 4.24VSYY96 pKa = 11.1LPYY99 pKa = 10.19EE100 pKa = 3.9QRR102 pKa = 11.84TYY104 pKa = 11.68SPLQLSKK111 pKa = 10.62GTVYY115 pKa = 10.8PPGKK119 pKa = 9.57EE120 pKa = 3.99VNNLFVTATTSTVFTTGIIRR140 pKa = 11.84EE141 pKa = 4.38HH142 pKa = 6.47PTDD145 pKa = 3.61ALYY148 pKa = 9.05VTYY151 pKa = 10.31EE152 pKa = 4.15GSTKK156 pKa = 10.25PGFSGVPYY164 pKa = 10.31VSNKK168 pKa = 9.52KK169 pKa = 9.74AVAMHH174 pKa = 6.89LGSNSYY180 pKa = 10.99GKK182 pKa = 10.25GIKK185 pKa = 9.98YY186 pKa = 9.97SWILIMIYY194 pKa = 9.68EE195 pKa = 4.5AEE197 pKa = 4.18NPNQNVRR204 pKa = 11.84HH205 pKa = 5.64YY206 pKa = 11.0SDD208 pKa = 3.18EE209 pKa = 4.24TEE211 pKa = 3.81KK212 pKa = 11.16VFIRR216 pKa = 11.84EE217 pKa = 4.22STGSDD222 pKa = 2.97TYY224 pKa = 11.52EE225 pKa = 4.03KK226 pKa = 10.72LIAQALEE233 pKa = 3.96RR234 pKa = 11.84MSQGEE239 pKa = 4.5RR240 pKa = 11.84YY241 pKa = 9.16FKK243 pKa = 10.18RR244 pKa = 11.84AGYY247 pKa = 10.23RR248 pKa = 11.84GMDD251 pKa = 2.85SYY253 pKa = 11.84LFNNSDD259 pKa = 3.18GTMMEE264 pKa = 4.28ITPHH268 pKa = 5.32TRR270 pKa = 11.84QHH272 pKa = 4.67MMAFNEE278 pKa = 4.01YY279 pKa = 10.92DD280 pKa = 3.27NDD282 pKa = 4.04AEE284 pKa = 4.31MMSGGYY290 pKa = 8.66RR291 pKa = 11.84RR292 pKa = 11.84KK293 pKa = 9.88RR294 pKa = 11.84SRR296 pKa = 11.84KK297 pKa = 9.12APSGWGEE304 pKa = 4.14LEE306 pKa = 3.6EE307 pKa = 4.21LAPVRR312 pKa = 11.84KK313 pKa = 10.06GKK315 pKa = 10.92GKK317 pKa = 10.39GGKK320 pKa = 8.6FRR322 pKa = 11.84YY323 pKa = 9.69DD324 pKa = 3.24EE325 pKa = 4.5PPAFKK330 pKa = 10.58GKK332 pKa = 9.89SWVDD336 pKa = 3.13MMEE339 pKa = 3.92EE340 pKa = 3.54SDD342 pKa = 4.24YY343 pKa = 11.68EE344 pKa = 4.17EE345 pKa = 4.94EE346 pKa = 4.72NADD349 pKa = 5.67DD350 pKa = 4.45SNQDD354 pKa = 3.02QPMVSQVKK362 pKa = 9.63NKK364 pKa = 10.21KK365 pKa = 9.7AIKK368 pKa = 10.21KK369 pKa = 10.06LGLDD373 pKa = 3.77NVPQTAMLVPDD384 pKa = 4.11ISRR387 pKa = 11.84PTVPVYY393 pKa = 10.75SDD395 pKa = 3.41YY396 pKa = 11.54PEE398 pKa = 4.03VVKK401 pKa = 10.09TSKK404 pKa = 10.83NVVSPPDD411 pKa = 3.25VEE413 pKa = 5.55APGKK417 pKa = 7.45TQTAEE422 pKa = 3.89EE423 pKa = 4.31VVIPLRR429 pKa = 11.84TTTGSLPTPNFNLDD443 pKa = 3.57HH444 pKa = 7.06LSNTAAPPLTLAQLGEE460 pKa = 4.38ALPLILEE467 pKa = 4.46SLNSLSNRR475 pKa = 11.84LQNPRR480 pKa = 11.84RR481 pKa = 11.84RR482 pKa = 11.84RR483 pKa = 11.84SRR485 pKa = 11.84KK486 pKa = 9.17PSAKK490 pKa = 9.65PSNSCAQEE498 pKa = 3.64
Molecular weight: 55.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
890 |
392 |
498 |
445.0 |
50.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.955 ± 0.222 | 1.348 ± 0.779 |
5.393 ± 0.307 | 6.629 ± 0.163 |
3.258 ± 0.533 | 5.955 ± 0.718 |
1.461 ± 0.285 | 4.831 ± 1.332 |
5.73 ± 0.242 | 7.64 ± 0.834 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.596 ± 0.315 | 4.27 ± 0.783 |
6.404 ± 0.678 | 4.27 ± 0.539 |
5.393 ± 0.023 | 6.854 ± 1.135 |
6.966 ± 0.712 | 7.191 ± 0.196 |
1.685 ± 0.561 | 5.169 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
