
Changjiang sobemo-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
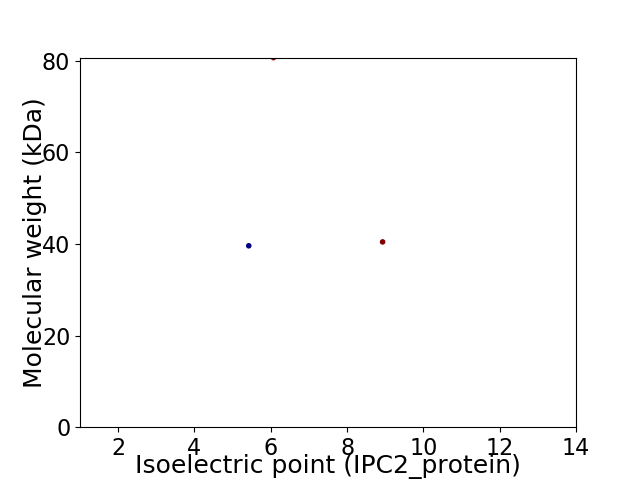
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
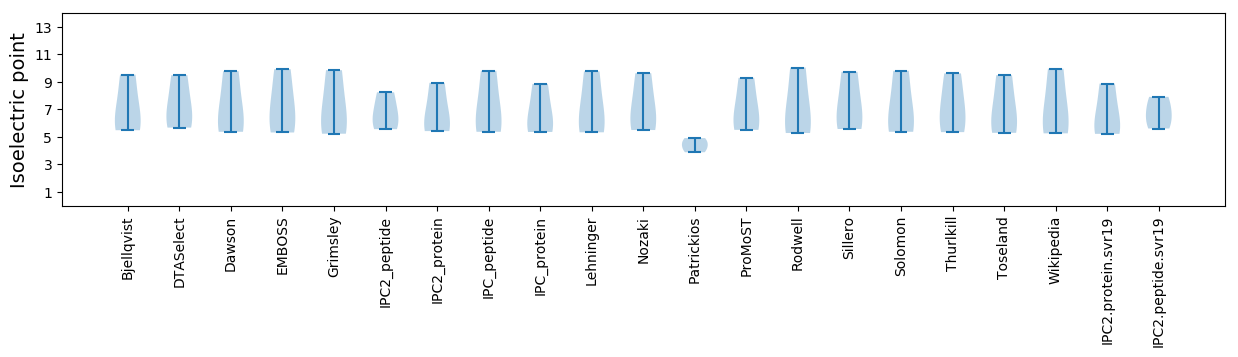
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KED0|A0A1L3KED0_9VIRU Uncharacterized protein OS=Changjiang sobemo-like virus 1 OX=1922800 PE=4 SV=1
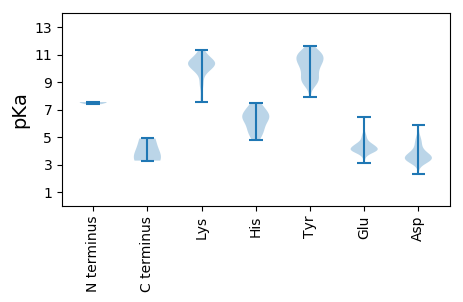
MM1 pKa = 7.51IRR3 pKa = 11.84EE4 pKa = 4.56AFAKK8 pKa = 10.01MNGTAVPGTPYY19 pKa = 11.23AFTYY23 pKa = 9.11PRR25 pKa = 11.84KK26 pKa = 9.94SDD28 pKa = 3.57WIEE31 pKa = 3.8KK32 pKa = 10.38DD33 pKa = 3.7LEE35 pKa = 4.38CLVDD39 pKa = 3.61MVLIRR44 pKa = 11.84LCLLEE49 pKa = 4.35YY50 pKa = 10.52SSSSTVEE57 pKa = 3.48ALYY60 pKa = 8.8NTTLIDD66 pKa = 4.91LGYY69 pKa = 9.51QDD71 pKa = 4.35PVSVFVKK78 pKa = 10.54DD79 pKa = 3.83EE80 pKa = 3.74PHH82 pKa = 6.35TIEE85 pKa = 5.14KK86 pKa = 9.88IKK88 pKa = 10.03QGRR91 pKa = 11.84YY92 pKa = 9.33RR93 pKa = 11.84LIANCTLVEE102 pKa = 4.0EE103 pKa = 4.91AIKK106 pKa = 10.63RR107 pKa = 11.84LLFAEE112 pKa = 4.55QNDD115 pKa = 3.71ADD117 pKa = 5.87KK118 pKa = 11.26NDD120 pKa = 4.38CYY122 pKa = 11.23NEE124 pKa = 4.35CGSAGGWDD132 pKa = 3.37WTTDD136 pKa = 3.08EE137 pKa = 4.05AAARR141 pKa = 11.84CFEE144 pKa = 5.21CIRR147 pKa = 11.84PWLPNAATSDD157 pKa = 3.44VSAWDD162 pKa = 3.48WSVKK166 pKa = 7.62EE167 pKa = 4.05WMFMAEE173 pKa = 3.79LEE175 pKa = 4.01IRR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84LNGASTKK186 pKa = 10.51SSWYY190 pKa = 8.9KK191 pKa = 10.08ISYY194 pKa = 9.24NVYY197 pKa = 9.86KK198 pKa = 10.26SFCRR202 pKa = 11.84SVYY205 pKa = 10.81VLTDD209 pKa = 3.52GSAYY213 pKa = 8.95TLEE216 pKa = 4.45HH217 pKa = 6.72PGVQKK222 pKa = 10.99SGDD225 pKa = 3.79YY226 pKa = 9.18NTTTTNGRR234 pKa = 11.84IRR236 pKa = 11.84NLIAVLVGSDD246 pKa = 3.26KK247 pKa = 11.32SKK249 pKa = 11.21SAGDD253 pKa = 4.06DD254 pKa = 4.21NISTWVDD261 pKa = 3.29DD262 pKa = 4.92AVQKK266 pKa = 8.56YY267 pKa = 9.19AKK269 pKa = 10.15YY270 pKa = 9.9GIRR273 pKa = 11.84VTDD276 pKa = 3.45YY277 pKa = 11.17EE278 pKa = 4.14RR279 pKa = 11.84VKK281 pKa = 10.79DD282 pKa = 3.99GKK284 pKa = 11.22VKK286 pKa = 10.41FCSATFTEE294 pKa = 5.26SKK296 pKa = 10.41AIPDD300 pKa = 3.58SAMKK304 pKa = 10.29SLYY307 pKa = 10.06KK308 pKa = 10.57LLWSQPSYY316 pKa = 10.89MLLDD320 pKa = 3.6TFRR323 pKa = 11.84NEE325 pKa = 3.6FRR327 pKa = 11.84HH328 pKa = 6.61APDD331 pKa = 3.85LEE333 pKa = 4.13RR334 pKa = 11.84AEE336 pKa = 4.26AAILRR341 pKa = 11.84SGYY344 pKa = 10.49LADD347 pKa = 4.35LAA349 pKa = 4.89
MM1 pKa = 7.51IRR3 pKa = 11.84EE4 pKa = 4.56AFAKK8 pKa = 10.01MNGTAVPGTPYY19 pKa = 11.23AFTYY23 pKa = 9.11PRR25 pKa = 11.84KK26 pKa = 9.94SDD28 pKa = 3.57WIEE31 pKa = 3.8KK32 pKa = 10.38DD33 pKa = 3.7LEE35 pKa = 4.38CLVDD39 pKa = 3.61MVLIRR44 pKa = 11.84LCLLEE49 pKa = 4.35YY50 pKa = 10.52SSSSTVEE57 pKa = 3.48ALYY60 pKa = 8.8NTTLIDD66 pKa = 4.91LGYY69 pKa = 9.51QDD71 pKa = 4.35PVSVFVKK78 pKa = 10.54DD79 pKa = 3.83EE80 pKa = 3.74PHH82 pKa = 6.35TIEE85 pKa = 5.14KK86 pKa = 9.88IKK88 pKa = 10.03QGRR91 pKa = 11.84YY92 pKa = 9.33RR93 pKa = 11.84LIANCTLVEE102 pKa = 4.0EE103 pKa = 4.91AIKK106 pKa = 10.63RR107 pKa = 11.84LLFAEE112 pKa = 4.55QNDD115 pKa = 3.71ADD117 pKa = 5.87KK118 pKa = 11.26NDD120 pKa = 4.38CYY122 pKa = 11.23NEE124 pKa = 4.35CGSAGGWDD132 pKa = 3.37WTTDD136 pKa = 3.08EE137 pKa = 4.05AAARR141 pKa = 11.84CFEE144 pKa = 5.21CIRR147 pKa = 11.84PWLPNAATSDD157 pKa = 3.44VSAWDD162 pKa = 3.48WSVKK166 pKa = 7.62EE167 pKa = 4.05WMFMAEE173 pKa = 3.79LEE175 pKa = 4.01IRR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84LNGASTKK186 pKa = 10.51SSWYY190 pKa = 8.9KK191 pKa = 10.08ISYY194 pKa = 9.24NVYY197 pKa = 9.86KK198 pKa = 10.26SFCRR202 pKa = 11.84SVYY205 pKa = 10.81VLTDD209 pKa = 3.52GSAYY213 pKa = 8.95TLEE216 pKa = 4.45HH217 pKa = 6.72PGVQKK222 pKa = 10.99SGDD225 pKa = 3.79YY226 pKa = 9.18NTTTTNGRR234 pKa = 11.84IRR236 pKa = 11.84NLIAVLVGSDD246 pKa = 3.26KK247 pKa = 11.32SKK249 pKa = 11.21SAGDD253 pKa = 4.06DD254 pKa = 4.21NISTWVDD261 pKa = 3.29DD262 pKa = 4.92AVQKK266 pKa = 8.56YY267 pKa = 9.19AKK269 pKa = 10.15YY270 pKa = 9.9GIRR273 pKa = 11.84VTDD276 pKa = 3.45YY277 pKa = 11.17EE278 pKa = 4.14RR279 pKa = 11.84VKK281 pKa = 10.79DD282 pKa = 3.99GKK284 pKa = 11.22VKK286 pKa = 10.41FCSATFTEE294 pKa = 5.26SKK296 pKa = 10.41AIPDD300 pKa = 3.58SAMKK304 pKa = 10.29SLYY307 pKa = 10.06KK308 pKa = 10.57LLWSQPSYY316 pKa = 10.89MLLDD320 pKa = 3.6TFRR323 pKa = 11.84NEE325 pKa = 3.6FRR327 pKa = 11.84HH328 pKa = 6.61APDD331 pKa = 3.85LEE333 pKa = 4.13RR334 pKa = 11.84AEE336 pKa = 4.26AAILRR341 pKa = 11.84SGYY344 pKa = 10.49LADD347 pKa = 4.35LAA349 pKa = 4.89
Molecular weight: 39.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KED0|A0A1L3KED0_9VIRU Uncharacterized protein OS=Changjiang sobemo-like virus 1 OX=1922800 PE=4 SV=1
MM1 pKa = 7.39RR2 pKa = 11.84QTMVKK7 pKa = 9.89KK8 pKa = 10.75VKK10 pKa = 10.32ANKK13 pKa = 9.65KK14 pKa = 9.05KK15 pKa = 10.3ALAVKK20 pKa = 10.32SSSSKK25 pKa = 9.64RR26 pKa = 11.84KK27 pKa = 7.94NRR29 pKa = 11.84RR30 pKa = 11.84MKK32 pKa = 9.97TKK34 pKa = 8.94MGIMRR39 pKa = 11.84TEE41 pKa = 4.09GKK43 pKa = 10.23LMKK46 pKa = 10.04AAQLIADD53 pKa = 4.68PCNGPLGYY61 pKa = 10.62GVGDD65 pKa = 4.71GYY67 pKa = 11.59NGAVSEE73 pKa = 4.31RR74 pKa = 11.84QRR76 pKa = 11.84TTIVSPSNNASNVSGYY92 pKa = 9.94IVWFPSYY99 pKa = 10.49HH100 pKa = 6.24GATGVGGNANLYY112 pKa = 9.32YY113 pKa = 10.66FEE115 pKa = 4.48TATPAVAPVNTIANPMGTTVNTSGTFLADD144 pKa = 3.39PTSASISAGGAFTRR158 pKa = 11.84AKK160 pKa = 10.12CLSACVQMAHH170 pKa = 7.03IGTLQTTAGQICTVRR185 pKa = 11.84NLSLAAFNTNYY196 pKa = 8.87GTVSPFQPMSVDD208 pKa = 3.27QLFGYY213 pKa = 9.97AHH215 pKa = 6.51EE216 pKa = 4.17RR217 pKa = 11.84QRR219 pKa = 11.84FNIDD223 pKa = 2.34GHH225 pKa = 4.81EE226 pKa = 4.5VVWRR230 pKa = 11.84PADD233 pKa = 3.4NSSVLRR239 pKa = 11.84SNGADD244 pKa = 3.42PAIATYY250 pKa = 10.29EE251 pKa = 4.01PDD253 pKa = 3.31VLFIQGNPTVNTTSSAVMDD272 pKa = 4.8PSSVMGIAIGFRR284 pKa = 11.84GLSPGLSSTMFNFVKK299 pKa = 10.56CSEE302 pKa = 4.63LEE304 pKa = 3.82LAPRR308 pKa = 11.84NNLMEE313 pKa = 4.53EE314 pKa = 4.21MPRR317 pKa = 11.84PVPGPRR323 pKa = 11.84YY324 pKa = 10.08SISQVTDD331 pKa = 3.22YY332 pKa = 11.02LDD334 pKa = 3.86SVDD337 pKa = 5.16PSWQSHH343 pKa = 5.47LMSKK347 pKa = 10.63LGGAATAAVKK357 pKa = 9.37TFAPTIVDD365 pKa = 3.41MFKK368 pKa = 10.06TKK370 pKa = 10.63LYY372 pKa = 10.43IKK374 pKa = 10.47DD375 pKa = 3.38GSLL378 pKa = 3.28
MM1 pKa = 7.39RR2 pKa = 11.84QTMVKK7 pKa = 9.89KK8 pKa = 10.75VKK10 pKa = 10.32ANKK13 pKa = 9.65KK14 pKa = 9.05KK15 pKa = 10.3ALAVKK20 pKa = 10.32SSSSKK25 pKa = 9.64RR26 pKa = 11.84KK27 pKa = 7.94NRR29 pKa = 11.84RR30 pKa = 11.84MKK32 pKa = 9.97TKK34 pKa = 8.94MGIMRR39 pKa = 11.84TEE41 pKa = 4.09GKK43 pKa = 10.23LMKK46 pKa = 10.04AAQLIADD53 pKa = 4.68PCNGPLGYY61 pKa = 10.62GVGDD65 pKa = 4.71GYY67 pKa = 11.59NGAVSEE73 pKa = 4.31RR74 pKa = 11.84QRR76 pKa = 11.84TTIVSPSNNASNVSGYY92 pKa = 9.94IVWFPSYY99 pKa = 10.49HH100 pKa = 6.24GATGVGGNANLYY112 pKa = 9.32YY113 pKa = 10.66FEE115 pKa = 4.48TATPAVAPVNTIANPMGTTVNTSGTFLADD144 pKa = 3.39PTSASISAGGAFTRR158 pKa = 11.84AKK160 pKa = 10.12CLSACVQMAHH170 pKa = 7.03IGTLQTTAGQICTVRR185 pKa = 11.84NLSLAAFNTNYY196 pKa = 8.87GTVSPFQPMSVDD208 pKa = 3.27QLFGYY213 pKa = 9.97AHH215 pKa = 6.51EE216 pKa = 4.17RR217 pKa = 11.84QRR219 pKa = 11.84FNIDD223 pKa = 2.34GHH225 pKa = 4.81EE226 pKa = 4.5VVWRR230 pKa = 11.84PADD233 pKa = 3.4NSSVLRR239 pKa = 11.84SNGADD244 pKa = 3.42PAIATYY250 pKa = 10.29EE251 pKa = 4.01PDD253 pKa = 3.31VLFIQGNPTVNTTSSAVMDD272 pKa = 4.8PSSVMGIAIGFRR284 pKa = 11.84GLSPGLSSTMFNFVKK299 pKa = 10.56CSEE302 pKa = 4.63LEE304 pKa = 3.82LAPRR308 pKa = 11.84NNLMEE313 pKa = 4.53EE314 pKa = 4.21MPRR317 pKa = 11.84PVPGPRR323 pKa = 11.84YY324 pKa = 10.08SISQVTDD331 pKa = 3.22YY332 pKa = 11.02LDD334 pKa = 3.86SVDD337 pKa = 5.16PSWQSHH343 pKa = 5.47LMSKK347 pKa = 10.63LGGAATAAVKK357 pKa = 9.37TFAPTIVDD365 pKa = 3.41MFKK368 pKa = 10.06TKK370 pKa = 10.63LYY372 pKa = 10.43IKK374 pKa = 10.47DD375 pKa = 3.38GSLL378 pKa = 3.28
Molecular weight: 40.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1449 |
349 |
722 |
483.0 |
53.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.248 ± 0.169 | 1.38 ± 0.357 |
5.176 ± 0.649 | 6.694 ± 1.394 |
3.244 ± 0.142 | 6.28 ± 0.659 |
1.035 ± 0.086 | 4.831 ± 0.193 |
7.039 ± 0.649 | 8.696 ± 0.92 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.692 ± 0.458 | 4.555 ± 0.458 |
4.762 ± 0.494 | 2.899 ± 0.33 |
5.176 ± 0.214 | 8.282 ± 0.414 |
5.728 ± 0.961 | 6.832 ± 0.28 |
1.311 ± 0.427 | 4.141 ± 0.385 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
