
Sewage-associated circular DNA virus-20
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
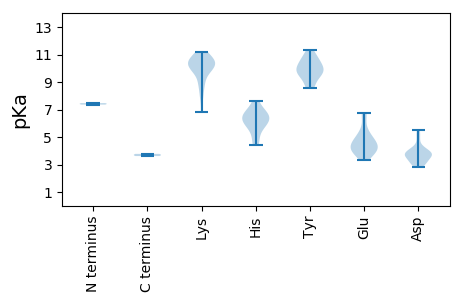
Average proteome isoelectric point is 7.42
Get precalculated fractions of proteins
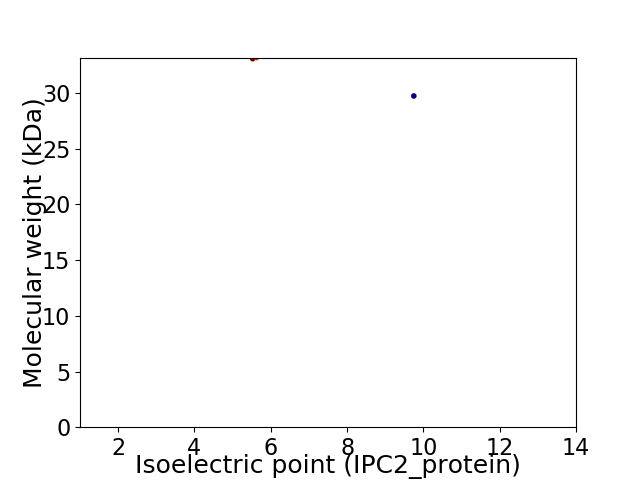
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UGX8|A0A0B4UGX8_9VIRU ATP-dependent helicase Rep OS=Sewage-associated circular DNA virus-20 OX=1592087 PE=3 SV=1
MM1 pKa = 7.36ARR3 pKa = 11.84SRR5 pKa = 11.84NYY7 pKa = 9.93VFTVNNYY14 pKa = 10.31DD15 pKa = 5.49DD16 pKa = 4.98EE17 pKa = 6.72DD18 pKa = 3.91EE19 pKa = 4.47HH20 pKa = 7.63QCWAMPWEE28 pKa = 4.42VKK30 pKa = 9.79DD31 pKa = 3.94CKK33 pKa = 10.45YY34 pKa = 10.53ICVGKK39 pKa = 10.38EE40 pKa = 3.35IGEE43 pKa = 4.38SGTPHH48 pKa = 5.82LQGYY52 pKa = 9.41ICFTQLKK59 pKa = 10.02SLNQLKK65 pKa = 10.63GFFPSAHH72 pKa = 6.48FEE74 pKa = 4.5TKK76 pKa = 10.35RR77 pKa = 11.84GTHH80 pKa = 4.91QQAADD85 pKa = 3.65YY86 pKa = 9.74CKK88 pKa = 10.45KK89 pKa = 10.82DD90 pKa = 2.87GDD92 pKa = 3.89FFEE95 pKa = 4.9WGTLPVDD102 pKa = 3.89DD103 pKa = 5.43CGSAGAAAMDD113 pKa = 4.15EE114 pKa = 4.52LMGHH118 pKa = 6.29TIEE121 pKa = 5.56CIRR124 pKa = 11.84NGDD127 pKa = 3.57YY128 pKa = 10.91KK129 pKa = 11.06GIPNAATHH137 pKa = 6.66FIKK140 pKa = 10.57AAEE143 pKa = 3.89YY144 pKa = 10.15RR145 pKa = 11.84VLKK148 pKa = 9.91EE149 pKa = 3.78QQQDD153 pKa = 3.25RR154 pKa = 11.84NLATLDD160 pKa = 3.7TLTHH164 pKa = 4.4QWRR167 pKa = 11.84WGKK170 pKa = 10.3AGCGKK175 pKa = 10.37SKK177 pKa = 9.85PARR180 pKa = 11.84DD181 pKa = 4.3ANPDD185 pKa = 3.51AYY187 pKa = 11.31LKK189 pKa = 9.85MCNKK193 pKa = 8.79WWDD196 pKa = 3.73GYY198 pKa = 9.11TGQEE202 pKa = 3.82VVIIEE207 pKa = 5.24DD208 pKa = 3.79FDD210 pKa = 4.9PDD212 pKa = 4.12HH213 pKa = 6.92KK214 pKa = 11.19CLVHH218 pKa = 6.82HH219 pKa = 6.39LKK221 pKa = 9.91IWADD225 pKa = 3.87RR226 pKa = 11.84YY227 pKa = 10.85AFPAEE232 pKa = 4.23VKK234 pKa = 10.3GGKK237 pKa = 8.98IDD239 pKa = 3.64IRR241 pKa = 11.84PKK243 pKa = 10.2TIIITSNYY251 pKa = 8.64HH252 pKa = 5.73PRR254 pKa = 11.84DD255 pKa = 3.34IFDD258 pKa = 4.36RR259 pKa = 11.84EE260 pKa = 3.97PDD262 pKa = 3.15LEE264 pKa = 6.56AIMRR268 pKa = 11.84RR269 pKa = 11.84FNVTHH274 pKa = 5.92VLGDD278 pKa = 3.79LGVDD282 pKa = 4.05FMQVDD287 pKa = 3.64NSS289 pKa = 3.62
MM1 pKa = 7.36ARR3 pKa = 11.84SRR5 pKa = 11.84NYY7 pKa = 9.93VFTVNNYY14 pKa = 10.31DD15 pKa = 5.49DD16 pKa = 4.98EE17 pKa = 6.72DD18 pKa = 3.91EE19 pKa = 4.47HH20 pKa = 7.63QCWAMPWEE28 pKa = 4.42VKK30 pKa = 9.79DD31 pKa = 3.94CKK33 pKa = 10.45YY34 pKa = 10.53ICVGKK39 pKa = 10.38EE40 pKa = 3.35IGEE43 pKa = 4.38SGTPHH48 pKa = 5.82LQGYY52 pKa = 9.41ICFTQLKK59 pKa = 10.02SLNQLKK65 pKa = 10.63GFFPSAHH72 pKa = 6.48FEE74 pKa = 4.5TKK76 pKa = 10.35RR77 pKa = 11.84GTHH80 pKa = 4.91QQAADD85 pKa = 3.65YY86 pKa = 9.74CKK88 pKa = 10.45KK89 pKa = 10.82DD90 pKa = 2.87GDD92 pKa = 3.89FFEE95 pKa = 4.9WGTLPVDD102 pKa = 3.89DD103 pKa = 5.43CGSAGAAAMDD113 pKa = 4.15EE114 pKa = 4.52LMGHH118 pKa = 6.29TIEE121 pKa = 5.56CIRR124 pKa = 11.84NGDD127 pKa = 3.57YY128 pKa = 10.91KK129 pKa = 11.06GIPNAATHH137 pKa = 6.66FIKK140 pKa = 10.57AAEE143 pKa = 3.89YY144 pKa = 10.15RR145 pKa = 11.84VLKK148 pKa = 9.91EE149 pKa = 3.78QQQDD153 pKa = 3.25RR154 pKa = 11.84NLATLDD160 pKa = 3.7TLTHH164 pKa = 4.4QWRR167 pKa = 11.84WGKK170 pKa = 10.3AGCGKK175 pKa = 10.37SKK177 pKa = 9.85PARR180 pKa = 11.84DD181 pKa = 4.3ANPDD185 pKa = 3.51AYY187 pKa = 11.31LKK189 pKa = 9.85MCNKK193 pKa = 8.79WWDD196 pKa = 3.73GYY198 pKa = 9.11TGQEE202 pKa = 3.82VVIIEE207 pKa = 5.24DD208 pKa = 3.79FDD210 pKa = 4.9PDD212 pKa = 4.12HH213 pKa = 6.92KK214 pKa = 11.19CLVHH218 pKa = 6.82HH219 pKa = 6.39LKK221 pKa = 9.91IWADD225 pKa = 3.87RR226 pKa = 11.84YY227 pKa = 10.85AFPAEE232 pKa = 4.23VKK234 pKa = 10.3GGKK237 pKa = 8.98IDD239 pKa = 3.64IRR241 pKa = 11.84PKK243 pKa = 10.2TIIITSNYY251 pKa = 8.64HH252 pKa = 5.73PRR254 pKa = 11.84DD255 pKa = 3.34IFDD258 pKa = 4.36RR259 pKa = 11.84EE260 pKa = 3.97PDD262 pKa = 3.15LEE264 pKa = 6.56AIMRR268 pKa = 11.84RR269 pKa = 11.84FNVTHH274 pKa = 5.92VLGDD278 pKa = 3.79LGVDD282 pKa = 4.05FMQVDD287 pKa = 3.64NSS289 pKa = 3.62
Molecular weight: 33.09 kDa
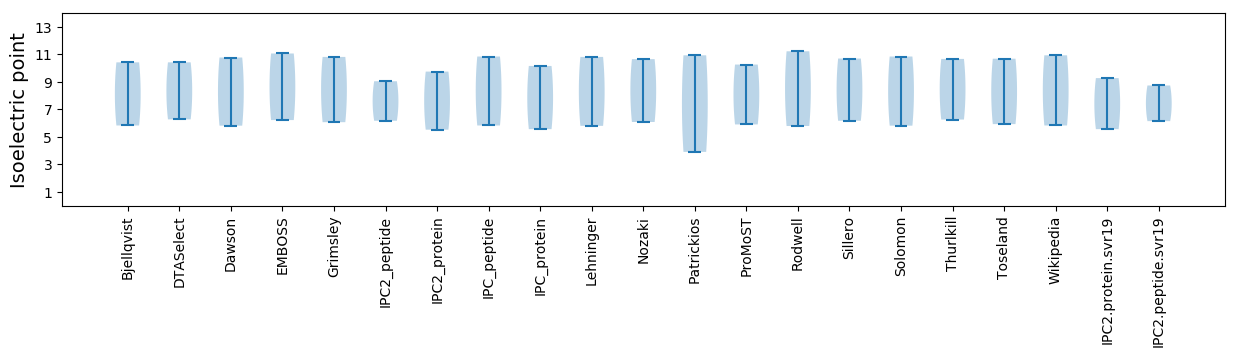
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGX8|A0A0B4UGX8_9VIRU ATP-dependent helicase Rep OS=Sewage-associated circular DNA virus-20 OX=1592087 PE=3 SV=1
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84KK4 pKa = 9.62SSKK7 pKa = 9.73ARR9 pKa = 11.84KK10 pKa = 7.96PLRR13 pKa = 11.84NNRR16 pKa = 11.84VKK18 pKa = 10.64RR19 pKa = 11.84AISRR23 pKa = 11.84KK24 pKa = 6.81SLRR27 pKa = 11.84RR28 pKa = 11.84QIATQIRR35 pKa = 11.84RR36 pKa = 11.84QAEE39 pKa = 4.06TKK41 pKa = 7.85EE42 pKa = 3.95QKK44 pKa = 8.46ITAEE48 pKa = 5.05RR49 pKa = 11.84YY50 pKa = 9.71LLWGPTAAAFNTRR63 pKa = 11.84NIFATSDD70 pKa = 3.01ILATISQGTGQGDD83 pKa = 3.76RR84 pKa = 11.84IGNKK88 pKa = 9.09VSLVNFFFSFIVWARR103 pKa = 11.84ATNLGGPSLPIDD115 pKa = 3.19LCMYY119 pKa = 9.62VFSDD123 pKa = 3.65KK124 pKa = 11.18LNPTSQLSLNIQNSIAGITQPYY146 pKa = 9.42FYY148 pKa = 10.08EE149 pKa = 4.75AGNATTGANLTITDD163 pKa = 3.17QLLRR167 pKa = 11.84VNGDD171 pKa = 2.8RR172 pKa = 11.84FTLHH176 pKa = 6.32KK177 pKa = 10.54KK178 pKa = 10.39KK179 pKa = 10.18IFKK182 pKa = 10.39ISTSTVANWSNNDD195 pKa = 3.09YY196 pKa = 10.51KK197 pKa = 11.05ASRR200 pKa = 11.84KK201 pKa = 9.66FKK203 pKa = 10.76INLAKK208 pKa = 10.66YY209 pKa = 8.59MPKK212 pKa = 10.16LVQWNDD218 pKa = 2.96AGVLTSRR225 pKa = 11.84QVWVMFLPVIANDD238 pKa = 3.54DD239 pKa = 3.82TSTVFANSDD248 pKa = 3.5LVGIGFDD255 pKa = 4.03YY256 pKa = 9.92QVKK259 pKa = 8.9WKK261 pKa = 11.0DD262 pKa = 3.19MM263 pKa = 3.78
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84KK4 pKa = 9.62SSKK7 pKa = 9.73ARR9 pKa = 11.84KK10 pKa = 7.96PLRR13 pKa = 11.84NNRR16 pKa = 11.84VKK18 pKa = 10.64RR19 pKa = 11.84AISRR23 pKa = 11.84KK24 pKa = 6.81SLRR27 pKa = 11.84RR28 pKa = 11.84QIATQIRR35 pKa = 11.84RR36 pKa = 11.84QAEE39 pKa = 4.06TKK41 pKa = 7.85EE42 pKa = 3.95QKK44 pKa = 8.46ITAEE48 pKa = 5.05RR49 pKa = 11.84YY50 pKa = 9.71LLWGPTAAAFNTRR63 pKa = 11.84NIFATSDD70 pKa = 3.01ILATISQGTGQGDD83 pKa = 3.76RR84 pKa = 11.84IGNKK88 pKa = 9.09VSLVNFFFSFIVWARR103 pKa = 11.84ATNLGGPSLPIDD115 pKa = 3.19LCMYY119 pKa = 9.62VFSDD123 pKa = 3.65KK124 pKa = 11.18LNPTSQLSLNIQNSIAGITQPYY146 pKa = 9.42FYY148 pKa = 10.08EE149 pKa = 4.75AGNATTGANLTITDD163 pKa = 3.17QLLRR167 pKa = 11.84VNGDD171 pKa = 2.8RR172 pKa = 11.84FTLHH176 pKa = 6.32KK177 pKa = 10.54KK178 pKa = 10.39KK179 pKa = 10.18IFKK182 pKa = 10.39ISTSTVANWSNNDD195 pKa = 3.09YY196 pKa = 10.51KK197 pKa = 11.05ASRR200 pKa = 11.84KK201 pKa = 9.66FKK203 pKa = 10.76INLAKK208 pKa = 10.66YY209 pKa = 8.59MPKK212 pKa = 10.16LVQWNDD218 pKa = 2.96AGVLTSRR225 pKa = 11.84QVWVMFLPVIANDD238 pKa = 3.54DD239 pKa = 3.82TSTVFANSDD248 pKa = 3.5LVGIGFDD255 pKa = 4.03YY256 pKa = 9.92QVKK259 pKa = 8.9WKK261 pKa = 11.0DD262 pKa = 3.19MM263 pKa = 3.78
Molecular weight: 29.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
552 |
263 |
289 |
276.0 |
31.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.971 ± 0.279 | 1.993 ± 1.141 |
7.246 ± 1.629 | 3.623 ± 1.487 |
4.891 ± 0.306 | 6.522 ± 0.848 |
2.355 ± 1.397 | 6.522 ± 0.497 |
7.428 ± 0.125 | 6.703 ± 0.907 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.174 ± 0.193 | 5.797 ± 1.279 |
3.623 ± 0.411 | 4.529 ± 0.293 |
5.978 ± 0.881 | 5.072 ± 1.791 |
6.522 ± 1.035 | 5.254 ± 0.318 |
2.536 ± 0.18 | 3.261 ± 0.424 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
