
Phomopsis vexans RNA virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus; unclassified Victorivirus
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
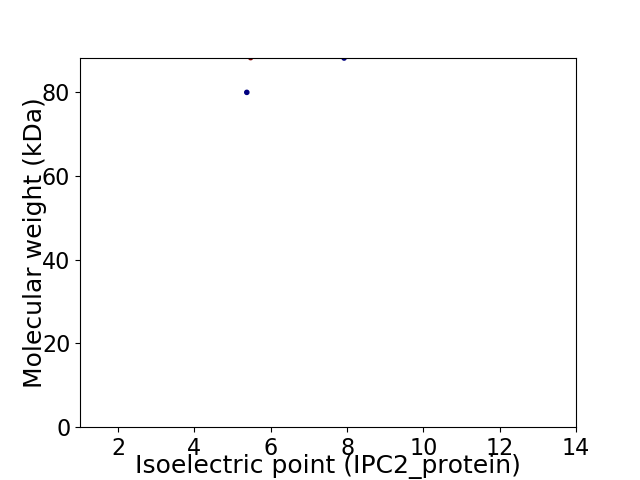
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
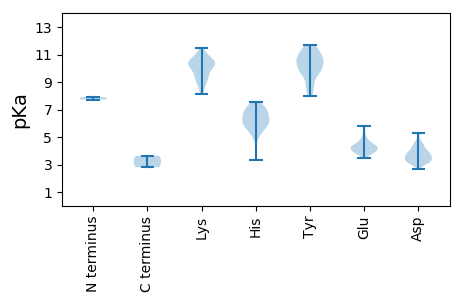
Protein with the lowest isoelectric point:
>tr|A0A0A7RU00|A0A0A7RU00_9VIRU Coat protein OS=Phomopsis vexans RNA virus OX=1580605 GN=CP PE=4 SV=1
MM1 pKa = 7.73ASTANFQAAVSSMLSGTLAGVPGGLLQQEE30 pKa = 4.14DD31 pKa = 4.5RR32 pKa = 11.84YY33 pKa = 10.63RR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 10.25RR37 pKa = 11.84SGLSIGVQEE46 pKa = 4.96HH47 pKa = 6.55GSLTYY52 pKa = 10.04SRR54 pKa = 11.84RR55 pKa = 11.84SIFYY59 pKa = 8.6EE60 pKa = 3.74VGRR63 pKa = 11.84RR64 pKa = 11.84TPRR67 pKa = 11.84LNDD70 pKa = 3.19ALAFTKK76 pKa = 10.44NDD78 pKa = 3.42EE79 pKa = 4.66EE80 pKa = 4.27ITMVDD85 pKa = 2.89ASVPINPAQAANFEE99 pKa = 4.05GWARR103 pKa = 11.84KK104 pKa = 9.44YY105 pKa = 11.34SNFSPQWEE113 pKa = 4.33MMDD116 pKa = 3.25LAGIVEE122 pKa = 4.63RR123 pKa = 11.84LAKK126 pKa = 10.16AVAAQSVYY134 pKa = 11.26GGVTCANLRR143 pKa = 11.84AGQPVRR149 pKa = 11.84VVALGTLDD157 pKa = 3.77SPQTASVNSVFIPRR171 pKa = 11.84TVEE174 pKa = 3.8TTTSDD179 pKa = 3.11GVFAVLSAAANGEE192 pKa = 4.36GASVTTDD199 pKa = 3.22VLRR202 pKa = 11.84LDD204 pKa = 4.51AGTNQPIVPTVDD216 pKa = 2.95NAVLATSCVEE226 pKa = 3.78ALRR229 pKa = 11.84ILGANLEE236 pKa = 4.19EE237 pKa = 4.99SGAGDD242 pKa = 3.6VFALAVTRR250 pKa = 11.84GVHH253 pKa = 5.6SVLSVVGHH261 pKa = 5.49TDD263 pKa = 2.69EE264 pKa = 5.28GGWIRR269 pKa = 11.84GVFRR273 pKa = 11.84HH274 pKa = 5.41CTFRR278 pKa = 11.84VPYY281 pKa = 10.51GGINPSLRR289 pKa = 11.84EE290 pKa = 4.08YY291 pKa = 10.56PGLPPLASTLTGHH304 pKa = 6.94ISAWCDD310 pKa = 3.48AIALKK315 pKa = 10.6SAAAVAHH322 pKa = 6.82CDD324 pKa = 3.38PCVTASGGTYY334 pKa = 7.97PTVFTSNRR342 pKa = 11.84GLVSPPGTDD351 pKa = 3.18EE352 pKa = 5.66ADD354 pKa = 4.17LGDD357 pKa = 4.47ADD359 pKa = 4.1ADD361 pKa = 3.71AVGRR365 pKa = 11.84QVGADD370 pKa = 3.71LGRR373 pKa = 11.84FSPVYY378 pKa = 9.63MDD380 pKa = 4.42ALTTIFGLRR389 pKa = 11.84AVSGVADD396 pKa = 3.5AVFRR400 pKa = 11.84TVGRR404 pKa = 11.84DD405 pKa = 3.73YY406 pKa = 11.7LNGAHH411 pKa = 7.13DD412 pKa = 3.22RR413 pKa = 11.84HH414 pKa = 6.18LRR416 pKa = 11.84HH417 pKa = 5.88KK418 pKa = 9.26TVAPYY423 pKa = 9.8FWIEE427 pKa = 3.9PTSLIPAGAFGTVAEE442 pKa = 4.28QAGFGALTTPGVEE455 pKa = 3.99RR456 pKa = 11.84EE457 pKa = 3.91QPAFEE462 pKa = 4.32KK463 pKa = 10.83VRR465 pKa = 11.84EE466 pKa = 4.05LDD468 pKa = 2.87HH469 pKa = 6.92GRR471 pKa = 11.84NANYY475 pKa = 8.85ATIAFKK481 pKa = 10.22MRR483 pKa = 11.84SARR486 pKa = 11.84TSGLICAYY494 pKa = 10.1AGTPASLTGLRR505 pKa = 11.84LYY507 pKa = 10.74QFDD510 pKa = 4.06EE511 pKa = 4.6EE512 pKa = 5.04SVVLAGDD519 pKa = 3.35QGPTGGRR526 pKa = 11.84VAQKK530 pKa = 10.12HH531 pKa = 5.12AAADD535 pKa = 4.59PISSYY540 pKa = 9.56LWRR543 pKa = 11.84RR544 pKa = 11.84GQSPIPAPAEE554 pKa = 3.84FVNTQGSYY562 pKa = 8.35AAKK565 pKa = 9.93YY566 pKa = 10.21KK567 pKa = 10.16IVEE570 pKa = 3.9WDD572 pKa = 3.31EE573 pKa = 5.79DD574 pKa = 3.81FDD576 pKa = 4.1ATLGDD581 pKa = 3.66LPEE584 pKa = 4.77AFEE587 pKa = 4.52LEE589 pKa = 4.48QYY591 pKa = 8.96SVRR594 pKa = 11.84WRR596 pKa = 11.84VTVPTGLPNGPSNAGDD612 pKa = 3.52SGAKK616 pKa = 9.33RR617 pKa = 11.84ARR619 pKa = 11.84SRR621 pKa = 11.84AAISLAQATIRR632 pKa = 11.84NRR634 pKa = 11.84GLGDD638 pKa = 3.55ANSPVISVSNVPPSWEE654 pKa = 4.3DD655 pKa = 3.41EE656 pKa = 4.35PGRR659 pKa = 11.84PMAADD664 pKa = 3.19IGDD667 pKa = 3.88HH668 pKa = 5.9RR669 pKa = 11.84VGVADD674 pKa = 3.93GQVYY678 pKa = 10.51ASGADD683 pKa = 3.13RR684 pKa = 11.84HH685 pKa = 5.95VRR687 pKa = 11.84GDD689 pKa = 3.6DD690 pKa = 3.1RR691 pKa = 11.84GAALNPVAHH700 pKa = 6.23HH701 pKa = 5.73QPLRR705 pKa = 11.84GAPYY709 pKa = 8.44PPRR712 pKa = 11.84GGGVLPGGGGPALPPQPPAGPPPAAPPSGGDD743 pKa = 3.09NGSDD747 pKa = 3.23VSGPANEE754 pKa = 4.36PDD756 pKa = 3.81AGNAAPAPPLL766 pKa = 3.63
MM1 pKa = 7.73ASTANFQAAVSSMLSGTLAGVPGGLLQQEE30 pKa = 4.14DD31 pKa = 4.5RR32 pKa = 11.84YY33 pKa = 10.63RR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 10.25RR37 pKa = 11.84SGLSIGVQEE46 pKa = 4.96HH47 pKa = 6.55GSLTYY52 pKa = 10.04SRR54 pKa = 11.84RR55 pKa = 11.84SIFYY59 pKa = 8.6EE60 pKa = 3.74VGRR63 pKa = 11.84RR64 pKa = 11.84TPRR67 pKa = 11.84LNDD70 pKa = 3.19ALAFTKK76 pKa = 10.44NDD78 pKa = 3.42EE79 pKa = 4.66EE80 pKa = 4.27ITMVDD85 pKa = 2.89ASVPINPAQAANFEE99 pKa = 4.05GWARR103 pKa = 11.84KK104 pKa = 9.44YY105 pKa = 11.34SNFSPQWEE113 pKa = 4.33MMDD116 pKa = 3.25LAGIVEE122 pKa = 4.63RR123 pKa = 11.84LAKK126 pKa = 10.16AVAAQSVYY134 pKa = 11.26GGVTCANLRR143 pKa = 11.84AGQPVRR149 pKa = 11.84VVALGTLDD157 pKa = 3.77SPQTASVNSVFIPRR171 pKa = 11.84TVEE174 pKa = 3.8TTTSDD179 pKa = 3.11GVFAVLSAAANGEE192 pKa = 4.36GASVTTDD199 pKa = 3.22VLRR202 pKa = 11.84LDD204 pKa = 4.51AGTNQPIVPTVDD216 pKa = 2.95NAVLATSCVEE226 pKa = 3.78ALRR229 pKa = 11.84ILGANLEE236 pKa = 4.19EE237 pKa = 4.99SGAGDD242 pKa = 3.6VFALAVTRR250 pKa = 11.84GVHH253 pKa = 5.6SVLSVVGHH261 pKa = 5.49TDD263 pKa = 2.69EE264 pKa = 5.28GGWIRR269 pKa = 11.84GVFRR273 pKa = 11.84HH274 pKa = 5.41CTFRR278 pKa = 11.84VPYY281 pKa = 10.51GGINPSLRR289 pKa = 11.84EE290 pKa = 4.08YY291 pKa = 10.56PGLPPLASTLTGHH304 pKa = 6.94ISAWCDD310 pKa = 3.48AIALKK315 pKa = 10.6SAAAVAHH322 pKa = 6.82CDD324 pKa = 3.38PCVTASGGTYY334 pKa = 7.97PTVFTSNRR342 pKa = 11.84GLVSPPGTDD351 pKa = 3.18EE352 pKa = 5.66ADD354 pKa = 4.17LGDD357 pKa = 4.47ADD359 pKa = 4.1ADD361 pKa = 3.71AVGRR365 pKa = 11.84QVGADD370 pKa = 3.71LGRR373 pKa = 11.84FSPVYY378 pKa = 9.63MDD380 pKa = 4.42ALTTIFGLRR389 pKa = 11.84AVSGVADD396 pKa = 3.5AVFRR400 pKa = 11.84TVGRR404 pKa = 11.84DD405 pKa = 3.73YY406 pKa = 11.7LNGAHH411 pKa = 7.13DD412 pKa = 3.22RR413 pKa = 11.84HH414 pKa = 6.18LRR416 pKa = 11.84HH417 pKa = 5.88KK418 pKa = 9.26TVAPYY423 pKa = 9.8FWIEE427 pKa = 3.9PTSLIPAGAFGTVAEE442 pKa = 4.28QAGFGALTTPGVEE455 pKa = 3.99RR456 pKa = 11.84EE457 pKa = 3.91QPAFEE462 pKa = 4.32KK463 pKa = 10.83VRR465 pKa = 11.84EE466 pKa = 4.05LDD468 pKa = 2.87HH469 pKa = 6.92GRR471 pKa = 11.84NANYY475 pKa = 8.85ATIAFKK481 pKa = 10.22MRR483 pKa = 11.84SARR486 pKa = 11.84TSGLICAYY494 pKa = 10.1AGTPASLTGLRR505 pKa = 11.84LYY507 pKa = 10.74QFDD510 pKa = 4.06EE511 pKa = 4.6EE512 pKa = 5.04SVVLAGDD519 pKa = 3.35QGPTGGRR526 pKa = 11.84VAQKK530 pKa = 10.12HH531 pKa = 5.12AAADD535 pKa = 4.59PISSYY540 pKa = 9.56LWRR543 pKa = 11.84RR544 pKa = 11.84GQSPIPAPAEE554 pKa = 3.84FVNTQGSYY562 pKa = 8.35AAKK565 pKa = 9.93YY566 pKa = 10.21KK567 pKa = 10.16IVEE570 pKa = 3.9WDD572 pKa = 3.31EE573 pKa = 5.79DD574 pKa = 3.81FDD576 pKa = 4.1ATLGDD581 pKa = 3.66LPEE584 pKa = 4.77AFEE587 pKa = 4.52LEE589 pKa = 4.48QYY591 pKa = 8.96SVRR594 pKa = 11.84WRR596 pKa = 11.84VTVPTGLPNGPSNAGDD612 pKa = 3.52SGAKK616 pKa = 9.33RR617 pKa = 11.84ARR619 pKa = 11.84SRR621 pKa = 11.84AAISLAQATIRR632 pKa = 11.84NRR634 pKa = 11.84GLGDD638 pKa = 3.55ANSPVISVSNVPPSWEE654 pKa = 4.3DD655 pKa = 3.41EE656 pKa = 4.35PGRR659 pKa = 11.84PMAADD664 pKa = 3.19IGDD667 pKa = 3.88HH668 pKa = 5.9RR669 pKa = 11.84VGVADD674 pKa = 3.93GQVYY678 pKa = 10.51ASGADD683 pKa = 3.13RR684 pKa = 11.84HH685 pKa = 5.95VRR687 pKa = 11.84GDD689 pKa = 3.6DD690 pKa = 3.1RR691 pKa = 11.84GAALNPVAHH700 pKa = 6.23HH701 pKa = 5.73QPLRR705 pKa = 11.84GAPYY709 pKa = 8.44PPRR712 pKa = 11.84GGGVLPGGGGPALPPQPPAGPPPAAPPSGGDD743 pKa = 3.09NGSDD747 pKa = 3.23VSGPANEE754 pKa = 4.36PDD756 pKa = 3.81AGNAAPAPPLL766 pKa = 3.63
Molecular weight: 80.01 kDa
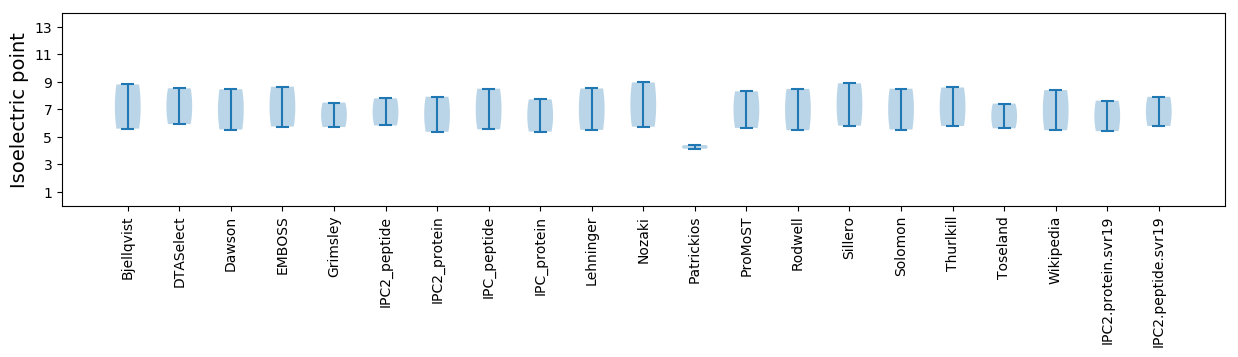
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7RU00|A0A0A7RU00_9VIRU Coat protein OS=Phomopsis vexans RNA virus OX=1580605 GN=CP PE=4 SV=1
MM1 pKa = 7.9PFAQQISFVYY11 pKa = 9.89RR12 pKa = 11.84PRR14 pKa = 11.84WNNTVPSRR22 pKa = 11.84LARR25 pKa = 11.84SAASFLHH32 pKa = 6.48CFVPVQVPLEE42 pKa = 4.04QGDD45 pKa = 4.51LDD47 pKa = 4.75LLTGQLCGHH56 pKa = 7.15LTMPPDD62 pKa = 3.81AGFSSMVVDD71 pKa = 4.43RR72 pKa = 11.84DD73 pKa = 3.05EE74 pKa = 5.7AMAAFPPKK82 pKa = 9.65NYY84 pKa = 10.81AMAIEE89 pKa = 4.28KK90 pKa = 10.84ANIYY94 pKa = 9.83VDD96 pKa = 4.42EE97 pKa = 4.88IARR100 pKa = 11.84DD101 pKa = 4.03MYY103 pKa = 11.27KK104 pKa = 8.68HH105 pKa = 6.08TPTEE109 pKa = 3.92LLRR112 pKa = 11.84AGPYY116 pKa = 9.96LRR118 pKa = 11.84RR119 pKa = 11.84LRR121 pKa = 11.84DD122 pKa = 3.09KK123 pKa = 11.27GVTHH127 pKa = 6.97DD128 pKa = 3.58QAAGFLLYY136 pKa = 10.61AATLSRR142 pKa = 11.84WCPHH146 pKa = 5.83AYY148 pKa = 9.16EE149 pKa = 3.82WAYY152 pKa = 10.93YY153 pKa = 10.64AITDD157 pKa = 3.83PAAAKK162 pKa = 9.8EE163 pKa = 4.07VSDD166 pKa = 3.73YY167 pKa = 11.48LKK169 pKa = 11.0AVGANSSSYY178 pKa = 10.89GALLVEE184 pKa = 4.5TDD186 pKa = 3.46TLQGRR191 pKa = 11.84GVPGTDD197 pKa = 4.26LPSDD201 pKa = 3.53SAKK204 pKa = 10.36RR205 pKa = 11.84CNLAALKK212 pKa = 10.52EE213 pKa = 4.14DD214 pKa = 3.96MLAEE218 pKa = 4.77LDD220 pKa = 4.01PVALRR225 pKa = 11.84SAVRR229 pKa = 11.84RR230 pKa = 11.84VLRR233 pKa = 11.84HH234 pKa = 5.34EE235 pKa = 3.98LRR237 pKa = 11.84HH238 pKa = 6.29RR239 pKa = 11.84DD240 pKa = 3.13GNYY243 pKa = 10.82ALDD246 pKa = 4.09FPTLEE251 pKa = 4.42EE252 pKa = 4.17HH253 pKa = 6.85WSSRR257 pKa = 11.84WSWAVNGSHH266 pKa = 6.71SSMVSKK272 pKa = 8.94TVPMVGVRR280 pKa = 11.84GKK282 pKa = 9.75HH283 pKa = 3.3VHH285 pKa = 6.09KK286 pKa = 9.74YY287 pKa = 9.53HH288 pKa = 6.37RR289 pKa = 11.84RR290 pKa = 11.84AWLEE294 pKa = 3.78SVRR297 pKa = 11.84DD298 pKa = 3.79DD299 pKa = 5.13PRR301 pKa = 11.84PNWEE305 pKa = 3.5GKK307 pKa = 8.14TFVSCNPKK315 pKa = 10.27LEE317 pKa = 4.43CGKK320 pKa = 8.26TRR322 pKa = 11.84AIYY325 pKa = 10.51ACDD328 pKa = 3.34TVSYY332 pKa = 10.56LAFEE336 pKa = 4.62HH337 pKa = 6.75LMAPVEE343 pKa = 4.23RR344 pKa = 11.84AWQGRR349 pKa = 11.84RR350 pKa = 11.84ILLNPGKK357 pKa = 10.35GGSIGLAEE365 pKa = 4.0RR366 pKa = 11.84VKK368 pKa = 10.44RR369 pKa = 11.84CRR371 pKa = 11.84DD372 pKa = 3.22RR373 pKa = 11.84SGVSMMLDD381 pKa = 3.24YY382 pKa = 11.46TDD384 pKa = 5.3FNSHH388 pKa = 5.96HH389 pKa = 5.94TTSAQQILIEE399 pKa = 4.33EE400 pKa = 4.54TCALTGYY407 pKa = 9.96PPDD410 pKa = 3.59LTAKK414 pKa = 10.23LVRR417 pKa = 11.84SFEE420 pKa = 4.07LQDD423 pKa = 3.24IYY425 pKa = 11.56LSGKK429 pKa = 9.8RR430 pKa = 11.84VGRR433 pKa = 11.84SAGTLMSGHH442 pKa = 7.01RR443 pKa = 11.84CTTYY447 pKa = 10.57INSVLNMAYY456 pKa = 10.81LMVVLGEE463 pKa = 4.27DD464 pKa = 3.87FVLEE468 pKa = 4.13RR469 pKa = 11.84QSLHH473 pKa = 7.28VGDD476 pKa = 5.16DD477 pKa = 3.73VYY479 pKa = 11.44LGARR483 pKa = 11.84DD484 pKa = 3.83YY485 pKa = 11.23PDD487 pKa = 4.04AGHH490 pKa = 7.54IITEE494 pKa = 4.28VMRR497 pKa = 11.84SPLRR501 pKa = 11.84MNKK504 pKa = 9.08SKK506 pKa = 10.94QSVGHH511 pKa = 6.22VGTEE515 pKa = 3.9LTRR518 pKa = 11.84VASNKK523 pKa = 9.33RR524 pKa = 11.84DD525 pKa = 3.47SFGYY529 pKa = 9.96LARR532 pKa = 11.84ATANLIAGNWYY543 pKa = 9.62SDD545 pKa = 3.35SVMDD549 pKa = 4.5PLEE552 pKa = 4.47GFTTMLAGCRR562 pKa = 11.84TLANRR567 pKa = 11.84GHH569 pKa = 6.15GMKK572 pKa = 10.49APLLLEE578 pKa = 4.26SAVKK582 pKa = 10.47RR583 pKa = 11.84ILGDD587 pKa = 3.7DD588 pKa = 4.22CPDD591 pKa = 3.7DD592 pKa = 4.15ALLRR596 pKa = 11.84RR597 pKa = 11.84ILLGDD602 pKa = 3.31IAVNSGPQYY611 pKa = 9.97SSSGRR616 pKa = 11.84HH617 pKa = 5.01VSIQVKK623 pKa = 10.77AEE625 pKa = 3.68FTTADD630 pKa = 3.47DD631 pKa = 3.98FGYY634 pKa = 11.06SLVPKK639 pKa = 10.27LATTTYY645 pKa = 9.63LSSHH649 pKa = 6.99ASPLEE654 pKa = 3.49IRR656 pKa = 11.84TLTEE660 pKa = 3.82CGIDD664 pKa = 3.54PSPSMVRR671 pKa = 11.84SSWKK675 pKa = 9.28KK676 pKa = 8.75SLRR679 pKa = 11.84IKK681 pKa = 10.32DD682 pKa = 3.41RR683 pKa = 11.84CLEE686 pKa = 3.98RR687 pKa = 11.84LVFGAVTTRR696 pKa = 11.84PARR699 pKa = 11.84GSIRR703 pKa = 11.84AEE705 pKa = 4.2DD706 pKa = 4.29LLKK709 pKa = 10.45QPKK712 pKa = 9.09PHH714 pKa = 6.55GCLLQFPLLVLAKK727 pKa = 10.51DD728 pKa = 4.66RR729 pKa = 11.84IPDD732 pKa = 3.58SEE734 pKa = 4.26LRR736 pKa = 11.84RR737 pKa = 11.84AVSDD741 pKa = 3.13SGGRR745 pKa = 11.84GWVEE749 pKa = 4.46DD750 pKa = 3.86IKK752 pKa = 11.45LEE754 pKa = 4.04AWGEE758 pKa = 3.99YY759 pKa = 8.43HH760 pKa = 7.53PSMIVNTGMSFSDD773 pKa = 3.33AAALSKK779 pKa = 10.43RR780 pKa = 11.84TDD782 pKa = 3.1ISVMTSTRR790 pKa = 11.84RR791 pKa = 11.84CYY793 pKa = 10.31VV794 pKa = 2.8
MM1 pKa = 7.9PFAQQISFVYY11 pKa = 9.89RR12 pKa = 11.84PRR14 pKa = 11.84WNNTVPSRR22 pKa = 11.84LARR25 pKa = 11.84SAASFLHH32 pKa = 6.48CFVPVQVPLEE42 pKa = 4.04QGDD45 pKa = 4.51LDD47 pKa = 4.75LLTGQLCGHH56 pKa = 7.15LTMPPDD62 pKa = 3.81AGFSSMVVDD71 pKa = 4.43RR72 pKa = 11.84DD73 pKa = 3.05EE74 pKa = 5.7AMAAFPPKK82 pKa = 9.65NYY84 pKa = 10.81AMAIEE89 pKa = 4.28KK90 pKa = 10.84ANIYY94 pKa = 9.83VDD96 pKa = 4.42EE97 pKa = 4.88IARR100 pKa = 11.84DD101 pKa = 4.03MYY103 pKa = 11.27KK104 pKa = 8.68HH105 pKa = 6.08TPTEE109 pKa = 3.92LLRR112 pKa = 11.84AGPYY116 pKa = 9.96LRR118 pKa = 11.84RR119 pKa = 11.84LRR121 pKa = 11.84DD122 pKa = 3.09KK123 pKa = 11.27GVTHH127 pKa = 6.97DD128 pKa = 3.58QAAGFLLYY136 pKa = 10.61AATLSRR142 pKa = 11.84WCPHH146 pKa = 5.83AYY148 pKa = 9.16EE149 pKa = 3.82WAYY152 pKa = 10.93YY153 pKa = 10.64AITDD157 pKa = 3.83PAAAKK162 pKa = 9.8EE163 pKa = 4.07VSDD166 pKa = 3.73YY167 pKa = 11.48LKK169 pKa = 11.0AVGANSSSYY178 pKa = 10.89GALLVEE184 pKa = 4.5TDD186 pKa = 3.46TLQGRR191 pKa = 11.84GVPGTDD197 pKa = 4.26LPSDD201 pKa = 3.53SAKK204 pKa = 10.36RR205 pKa = 11.84CNLAALKK212 pKa = 10.52EE213 pKa = 4.14DD214 pKa = 3.96MLAEE218 pKa = 4.77LDD220 pKa = 4.01PVALRR225 pKa = 11.84SAVRR229 pKa = 11.84RR230 pKa = 11.84VLRR233 pKa = 11.84HH234 pKa = 5.34EE235 pKa = 3.98LRR237 pKa = 11.84HH238 pKa = 6.29RR239 pKa = 11.84DD240 pKa = 3.13GNYY243 pKa = 10.82ALDD246 pKa = 4.09FPTLEE251 pKa = 4.42EE252 pKa = 4.17HH253 pKa = 6.85WSSRR257 pKa = 11.84WSWAVNGSHH266 pKa = 6.71SSMVSKK272 pKa = 8.94TVPMVGVRR280 pKa = 11.84GKK282 pKa = 9.75HH283 pKa = 3.3VHH285 pKa = 6.09KK286 pKa = 9.74YY287 pKa = 9.53HH288 pKa = 6.37RR289 pKa = 11.84RR290 pKa = 11.84AWLEE294 pKa = 3.78SVRR297 pKa = 11.84DD298 pKa = 3.79DD299 pKa = 5.13PRR301 pKa = 11.84PNWEE305 pKa = 3.5GKK307 pKa = 8.14TFVSCNPKK315 pKa = 10.27LEE317 pKa = 4.43CGKK320 pKa = 8.26TRR322 pKa = 11.84AIYY325 pKa = 10.51ACDD328 pKa = 3.34TVSYY332 pKa = 10.56LAFEE336 pKa = 4.62HH337 pKa = 6.75LMAPVEE343 pKa = 4.23RR344 pKa = 11.84AWQGRR349 pKa = 11.84RR350 pKa = 11.84ILLNPGKK357 pKa = 10.35GGSIGLAEE365 pKa = 4.0RR366 pKa = 11.84VKK368 pKa = 10.44RR369 pKa = 11.84CRR371 pKa = 11.84DD372 pKa = 3.22RR373 pKa = 11.84SGVSMMLDD381 pKa = 3.24YY382 pKa = 11.46TDD384 pKa = 5.3FNSHH388 pKa = 5.96HH389 pKa = 5.94TTSAQQILIEE399 pKa = 4.33EE400 pKa = 4.54TCALTGYY407 pKa = 9.96PPDD410 pKa = 3.59LTAKK414 pKa = 10.23LVRR417 pKa = 11.84SFEE420 pKa = 4.07LQDD423 pKa = 3.24IYY425 pKa = 11.56LSGKK429 pKa = 9.8RR430 pKa = 11.84VGRR433 pKa = 11.84SAGTLMSGHH442 pKa = 7.01RR443 pKa = 11.84CTTYY447 pKa = 10.57INSVLNMAYY456 pKa = 10.81LMVVLGEE463 pKa = 4.27DD464 pKa = 3.87FVLEE468 pKa = 4.13RR469 pKa = 11.84QSLHH473 pKa = 7.28VGDD476 pKa = 5.16DD477 pKa = 3.73VYY479 pKa = 11.44LGARR483 pKa = 11.84DD484 pKa = 3.83YY485 pKa = 11.23PDD487 pKa = 4.04AGHH490 pKa = 7.54IITEE494 pKa = 4.28VMRR497 pKa = 11.84SPLRR501 pKa = 11.84MNKK504 pKa = 9.08SKK506 pKa = 10.94QSVGHH511 pKa = 6.22VGTEE515 pKa = 3.9LTRR518 pKa = 11.84VASNKK523 pKa = 9.33RR524 pKa = 11.84DD525 pKa = 3.47SFGYY529 pKa = 9.96LARR532 pKa = 11.84ATANLIAGNWYY543 pKa = 9.62SDD545 pKa = 3.35SVMDD549 pKa = 4.5PLEE552 pKa = 4.47GFTTMLAGCRR562 pKa = 11.84TLANRR567 pKa = 11.84GHH569 pKa = 6.15GMKK572 pKa = 10.49APLLLEE578 pKa = 4.26SAVKK582 pKa = 10.47RR583 pKa = 11.84ILGDD587 pKa = 3.7DD588 pKa = 4.22CPDD591 pKa = 3.7DD592 pKa = 4.15ALLRR596 pKa = 11.84RR597 pKa = 11.84ILLGDD602 pKa = 3.31IAVNSGPQYY611 pKa = 9.97SSSGRR616 pKa = 11.84HH617 pKa = 5.01VSIQVKK623 pKa = 10.77AEE625 pKa = 3.68FTTADD630 pKa = 3.47DD631 pKa = 3.98FGYY634 pKa = 11.06SLVPKK639 pKa = 10.27LATTTYY645 pKa = 9.63LSSHH649 pKa = 6.99ASPLEE654 pKa = 3.49IRR656 pKa = 11.84TLTEE660 pKa = 3.82CGIDD664 pKa = 3.54PSPSMVRR671 pKa = 11.84SSWKK675 pKa = 9.28KK676 pKa = 8.75SLRR679 pKa = 11.84IKK681 pKa = 10.32DD682 pKa = 3.41RR683 pKa = 11.84CLEE686 pKa = 3.98RR687 pKa = 11.84LVFGAVTTRR696 pKa = 11.84PARR699 pKa = 11.84GSIRR703 pKa = 11.84AEE705 pKa = 4.2DD706 pKa = 4.29LLKK709 pKa = 10.45QPKK712 pKa = 9.09PHH714 pKa = 6.55GCLLQFPLLVLAKK727 pKa = 10.51DD728 pKa = 4.66RR729 pKa = 11.84IPDD732 pKa = 3.58SEE734 pKa = 4.26LRR736 pKa = 11.84RR737 pKa = 11.84AVSDD741 pKa = 3.13SGGRR745 pKa = 11.84GWVEE749 pKa = 4.46DD750 pKa = 3.86IKK752 pKa = 11.45LEE754 pKa = 4.04AWGEE758 pKa = 3.99YY759 pKa = 8.43HH760 pKa = 7.53PSMIVNTGMSFSDD773 pKa = 3.33AAALSKK779 pKa = 10.43RR780 pKa = 11.84TDD782 pKa = 3.1ISVMTSTRR790 pKa = 11.84RR791 pKa = 11.84CYY793 pKa = 10.31VV794 pKa = 2.8
Molecular weight: 88.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1560 |
766 |
794 |
780.0 |
84.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.282 ± 1.53 | 1.474 ± 0.374 |
6.026 ± 0.188 | 4.487 ± 0.119 |
2.756 ± 0.164 | 8.974 ± 1.414 |
2.5 ± 0.361 | 3.269 ± 0.091 |
2.821 ± 0.923 | 8.782 ± 1.155 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.051 ± 0.671 | 3.077 ± 0.298 |
6.538 ± 0.95 | 2.564 ± 0.292 |
7.5 ± 0.474 | 7.756 ± 0.471 |
5.833 ± 0.115 | 7.692 ± 0.529 |
1.41 ± 0.157 | 3.205 ± 0.309 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
