
Chitinophaga sp. MD30
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Chitinophaga; unclassified Chitinophaga
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
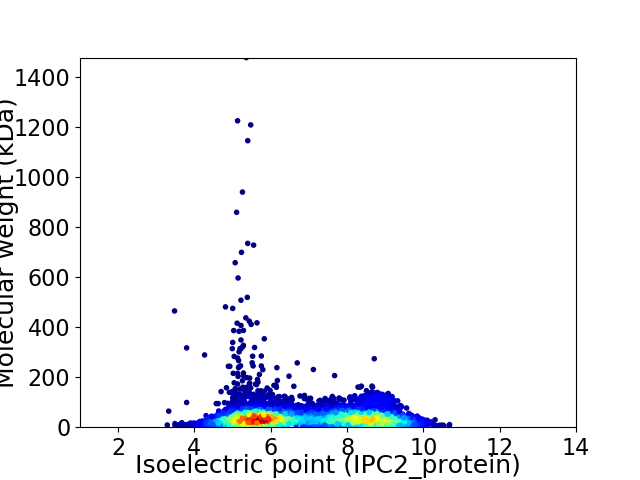
Virtual 2D-PAGE plot for 5689 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
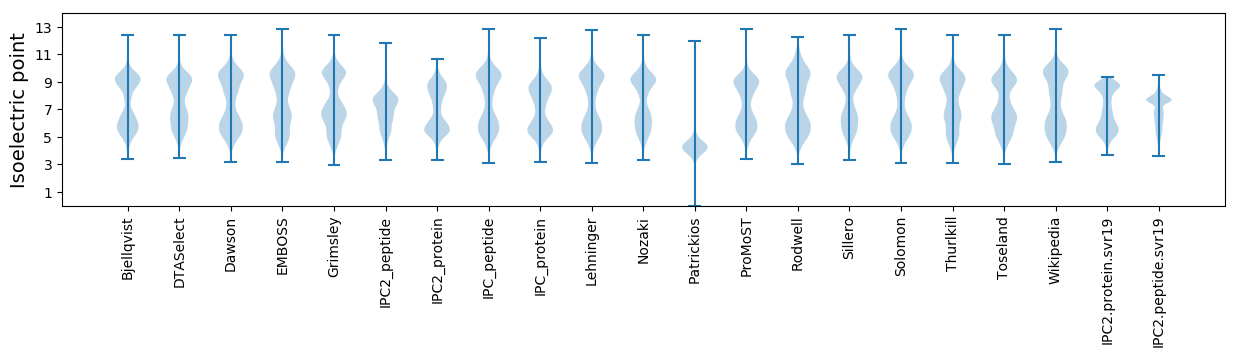
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A249T0A3|A0A249T0A3_9BACT NigD_N domain-containing protein OS=Chitinophaga sp. MD30 OX=2033437 GN=CK934_21350 PE=4 SV=1
MM1 pKa = 6.89VASLVTAPVNGTVVLNANGSFTYY24 pKa = 10.1TPNEE28 pKa = 4.05NFNGTDD34 pKa = 2.94SLTYY38 pKa = 8.76QVCDD42 pKa = 2.98NGTPSLCDD50 pKa = 2.92TATVVFNVAAINDD63 pKa = 3.78APVAVRR69 pKa = 11.84DD70 pKa = 4.21TVAVTEE76 pKa = 4.69DD77 pKa = 3.71LPATGNVLTNDD88 pKa = 3.74SDD90 pKa = 4.48PEE92 pKa = 5.26GDD94 pKa = 3.86ALVASLVTAPVNGTVVLNANGSFTYY119 pKa = 10.26IPNANFNGTDD129 pKa = 3.07SLTYY133 pKa = 8.76QVCDD137 pKa = 2.98NGTPSLCDD145 pKa = 2.92TATVVFNVAAINDD158 pKa = 3.78APVAVRR164 pKa = 11.84DD165 pKa = 4.21TVAVTEE171 pKa = 4.61DD172 pKa = 3.44VPATGNVLTNDD183 pKa = 3.74SDD185 pKa = 4.48PEE187 pKa = 5.26GDD189 pKa = 3.86ALVASLVTAPVNGTVVLNANGSFTYY214 pKa = 10.26IPNANFNGTDD224 pKa = 3.07SLTYY228 pKa = 8.76QVCDD232 pKa = 2.98NGTPSLCDD240 pKa = 2.92TATVVFNVAAINDD253 pKa = 3.78APVAVRR259 pKa = 11.84DD260 pKa = 4.21TVAVTEE266 pKa = 4.61DD267 pKa = 3.44VPATGNVLTNDD278 pKa = 3.74SDD280 pKa = 4.48PEE282 pKa = 5.26GDD284 pKa = 3.86ALVASLVTAPVNGTVVLNANGSFTYY309 pKa = 10.1TPNEE313 pKa = 4.05NFNGTDD319 pKa = 2.94SLTYY323 pKa = 8.76QVCDD327 pKa = 2.98NGTPSLCDD335 pKa = 2.92TATVVFNIAAVNDD348 pKa = 3.7APVAVRR354 pKa = 11.84DD355 pKa = 3.97TFQTRR360 pKa = 11.84QTIPVSGNVLTNDD373 pKa = 3.52TDD375 pKa = 4.03PEE377 pKa = 4.36GNALSTNVITGPLHH391 pKa = 5.07GTLVLNADD399 pKa = 3.59GSFIYY404 pKa = 9.97TPNAAFSGIDD414 pKa = 3.13QALYY418 pKa = 9.57RR419 pKa = 11.84VCDD422 pKa = 3.47NGVPSLCDD430 pKa = 2.68TATIVFVVTAVNTAPVAVNDD450 pKa = 3.67SFRR453 pKa = 11.84TVMQVPVSGNVLSNDD468 pKa = 3.58IDD470 pKa = 3.8PQGNGLQVALVDD482 pKa = 3.84NVKK485 pKa = 10.5QGSLQLNADD494 pKa = 3.21GTFRR498 pKa = 11.84YY499 pKa = 9.71EE500 pKa = 3.74PTGTFTGIDD509 pKa = 3.33SFVYY513 pKa = 9.89RR514 pKa = 11.84ICTPGASGLCDD525 pKa = 3.1SATVHH530 pKa = 5.8IQVTGVSEE538 pKa = 4.03QPMIGAALSVSTPVLQPDD556 pKa = 3.78GSYY559 pKa = 10.36RR560 pKa = 11.84VTYY563 pKa = 10.26LLRR566 pKa = 11.84IRR568 pKa = 11.84NYY570 pKa = 10.3GNVALRR576 pKa = 11.84QIEE579 pKa = 4.22ATDD582 pKa = 3.56DD583 pKa = 3.65LSRR586 pKa = 11.84VFPAPAGFRR595 pKa = 11.84VVSITADD602 pKa = 3.15GGLVANGTFDD612 pKa = 3.65GRR614 pKa = 11.84NDD616 pKa = 3.19INLLLSSSEE625 pKa = 3.82LALNVTSAISLVVDD639 pKa = 4.8IIPNGSFGPFNNNVTVQGSSLSGTIVTDD667 pKa = 3.49VSTNGSNPDD676 pKa = 3.34PSGNNDD682 pKa = 3.35PKK684 pKa = 11.32DD685 pKa = 3.72PGEE688 pKa = 4.27NLPTPLVLQPKK699 pKa = 9.76AIAGLAKK706 pKa = 10.44AATAPTAEE714 pKa = 3.69ISGNYY719 pKa = 8.0VVTYY723 pKa = 8.34TFVVKK728 pKa = 10.88NFGNVALNNVQVTDD742 pKa = 4.14DD743 pKa = 3.71LSKK746 pKa = 11.06VFSSPASFQIVGDD759 pKa = 3.4IRR761 pKa = 11.84TTGGLVGNNQYY772 pKa = 11.43NGINNTNLLSAGSGVNAGAADD793 pKa = 4.72TIQVTVRR800 pKa = 11.84IIPNKK805 pKa = 10.11QFGTFNNSAVLTGAAAGMGTTLTDD829 pKa = 4.29LSTNGLNADD838 pKa = 3.72PDD840 pKa = 4.48GNGVPDD846 pKa = 3.95EE847 pKa = 4.18QVVTPLLLNPTRR859 pKa = 11.84LRR861 pKa = 11.84IPEE864 pKa = 4.33GFSPNGDD871 pKa = 3.71GINDD875 pKa = 3.57RR876 pKa = 11.84FIIANAGNDD885 pKa = 3.78KK886 pKa = 10.67IQLEE890 pKa = 4.87VYY892 pKa = 9.8NRR894 pKa = 11.84WGNLVYY900 pKa = 11.02KK901 pKa = 10.21NVNYY905 pKa = 10.32RR906 pKa = 11.84NEE908 pKa = 4.0WDD910 pKa = 3.97GKK912 pKa = 9.69CNTGIHH918 pKa = 6.03IKK920 pKa = 10.58EE921 pKa = 5.09DD922 pKa = 3.7IPDD925 pKa = 3.4GTYY928 pKa = 10.82YY929 pKa = 11.01YY930 pKa = 10.07IVTATGSTGTEE941 pKa = 3.62RR942 pKa = 11.84YY943 pKa = 9.64VNYY946 pKa = 9.04ITIIRR951 pKa = 4.06
MM1 pKa = 6.89VASLVTAPVNGTVVLNANGSFTYY24 pKa = 10.1TPNEE28 pKa = 4.05NFNGTDD34 pKa = 2.94SLTYY38 pKa = 8.76QVCDD42 pKa = 2.98NGTPSLCDD50 pKa = 2.92TATVVFNVAAINDD63 pKa = 3.78APVAVRR69 pKa = 11.84DD70 pKa = 4.21TVAVTEE76 pKa = 4.69DD77 pKa = 3.71LPATGNVLTNDD88 pKa = 3.74SDD90 pKa = 4.48PEE92 pKa = 5.26GDD94 pKa = 3.86ALVASLVTAPVNGTVVLNANGSFTYY119 pKa = 10.26IPNANFNGTDD129 pKa = 3.07SLTYY133 pKa = 8.76QVCDD137 pKa = 2.98NGTPSLCDD145 pKa = 2.92TATVVFNVAAINDD158 pKa = 3.78APVAVRR164 pKa = 11.84DD165 pKa = 4.21TVAVTEE171 pKa = 4.61DD172 pKa = 3.44VPATGNVLTNDD183 pKa = 3.74SDD185 pKa = 4.48PEE187 pKa = 5.26GDD189 pKa = 3.86ALVASLVTAPVNGTVVLNANGSFTYY214 pKa = 10.26IPNANFNGTDD224 pKa = 3.07SLTYY228 pKa = 8.76QVCDD232 pKa = 2.98NGTPSLCDD240 pKa = 2.92TATVVFNVAAINDD253 pKa = 3.78APVAVRR259 pKa = 11.84DD260 pKa = 4.21TVAVTEE266 pKa = 4.61DD267 pKa = 3.44VPATGNVLTNDD278 pKa = 3.74SDD280 pKa = 4.48PEE282 pKa = 5.26GDD284 pKa = 3.86ALVASLVTAPVNGTVVLNANGSFTYY309 pKa = 10.1TPNEE313 pKa = 4.05NFNGTDD319 pKa = 2.94SLTYY323 pKa = 8.76QVCDD327 pKa = 2.98NGTPSLCDD335 pKa = 2.92TATVVFNIAAVNDD348 pKa = 3.7APVAVRR354 pKa = 11.84DD355 pKa = 3.97TFQTRR360 pKa = 11.84QTIPVSGNVLTNDD373 pKa = 3.52TDD375 pKa = 4.03PEE377 pKa = 4.36GNALSTNVITGPLHH391 pKa = 5.07GTLVLNADD399 pKa = 3.59GSFIYY404 pKa = 9.97TPNAAFSGIDD414 pKa = 3.13QALYY418 pKa = 9.57RR419 pKa = 11.84VCDD422 pKa = 3.47NGVPSLCDD430 pKa = 2.68TATIVFVVTAVNTAPVAVNDD450 pKa = 3.67SFRR453 pKa = 11.84TVMQVPVSGNVLSNDD468 pKa = 3.58IDD470 pKa = 3.8PQGNGLQVALVDD482 pKa = 3.84NVKK485 pKa = 10.5QGSLQLNADD494 pKa = 3.21GTFRR498 pKa = 11.84YY499 pKa = 9.71EE500 pKa = 3.74PTGTFTGIDD509 pKa = 3.33SFVYY513 pKa = 9.89RR514 pKa = 11.84ICTPGASGLCDD525 pKa = 3.1SATVHH530 pKa = 5.8IQVTGVSEE538 pKa = 4.03QPMIGAALSVSTPVLQPDD556 pKa = 3.78GSYY559 pKa = 10.36RR560 pKa = 11.84VTYY563 pKa = 10.26LLRR566 pKa = 11.84IRR568 pKa = 11.84NYY570 pKa = 10.3GNVALRR576 pKa = 11.84QIEE579 pKa = 4.22ATDD582 pKa = 3.56DD583 pKa = 3.65LSRR586 pKa = 11.84VFPAPAGFRR595 pKa = 11.84VVSITADD602 pKa = 3.15GGLVANGTFDD612 pKa = 3.65GRR614 pKa = 11.84NDD616 pKa = 3.19INLLLSSSEE625 pKa = 3.82LALNVTSAISLVVDD639 pKa = 4.8IIPNGSFGPFNNNVTVQGSSLSGTIVTDD667 pKa = 3.49VSTNGSNPDD676 pKa = 3.34PSGNNDD682 pKa = 3.35PKK684 pKa = 11.32DD685 pKa = 3.72PGEE688 pKa = 4.27NLPTPLVLQPKK699 pKa = 9.76AIAGLAKK706 pKa = 10.44AATAPTAEE714 pKa = 3.69ISGNYY719 pKa = 8.0VVTYY723 pKa = 8.34TFVVKK728 pKa = 10.88NFGNVALNNVQVTDD742 pKa = 4.14DD743 pKa = 3.71LSKK746 pKa = 11.06VFSSPASFQIVGDD759 pKa = 3.4IRR761 pKa = 11.84TTGGLVGNNQYY772 pKa = 11.43NGINNTNLLSAGSGVNAGAADD793 pKa = 4.72TIQVTVRR800 pKa = 11.84IIPNKK805 pKa = 10.11QFGTFNNSAVLTGAAAGMGTTLTDD829 pKa = 4.29LSTNGLNADD838 pKa = 3.72PDD840 pKa = 4.48GNGVPDD846 pKa = 3.95EE847 pKa = 4.18QVVTPLLLNPTRR859 pKa = 11.84LRR861 pKa = 11.84IPEE864 pKa = 4.33GFSPNGDD871 pKa = 3.71GINDD875 pKa = 3.57RR876 pKa = 11.84FIIANAGNDD885 pKa = 3.78KK886 pKa = 10.67IQLEE890 pKa = 4.87VYY892 pKa = 9.8NRR894 pKa = 11.84WGNLVYY900 pKa = 11.02KK901 pKa = 10.21NVNYY905 pKa = 10.32RR906 pKa = 11.84NEE908 pKa = 4.0WDD910 pKa = 3.97GKK912 pKa = 9.69CNTGIHH918 pKa = 6.03IKK920 pKa = 10.58EE921 pKa = 5.09DD922 pKa = 3.7IPDD925 pKa = 3.4GTYY928 pKa = 10.82YY929 pKa = 11.01YY930 pKa = 10.07IVTATGSTGTEE941 pKa = 3.62RR942 pKa = 11.84YY943 pKa = 9.64VNYY946 pKa = 9.04ITIIRR951 pKa = 4.06
Molecular weight: 98.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A249T3H9|A0A249T3H9_9BACT Penicillin-insensitive transglycosylase OS=Chitinophaga sp. MD30 OX=2033437 GN=CK934_27570 PE=4 SV=1
MM1 pKa = 7.56NIITSLTYY9 pKa = 10.76LLPGLLLGFLGSFHH23 pKa = 6.9CVGMCGPIALTLPVIHH39 pKa = 6.91LSSTRR44 pKa = 11.84KK45 pKa = 9.34LAGILLYY52 pKa = 10.72NAGRR56 pKa = 11.84TTTYY60 pKa = 11.13LLLGSLSGWLGQRR73 pKa = 11.84FFIGGLQQWLSILLGILLLLLLCWQYY99 pKa = 11.62ILQKK103 pKa = 9.68PLPQIPFFQQHH114 pKa = 5.07VKK116 pKa = 10.4SALGRR121 pKa = 11.84LLTRR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84FSTLFTIGFLNGLLPCGLVYY147 pKa = 10.62IGITGAIATGSAWKK161 pKa = 10.01GALFMTAFGIGTIPAMAAVSWFSQLLRR188 pKa = 11.84VHH190 pKa = 6.66IRR192 pKa = 11.84RR193 pKa = 11.84FIPVAIGMVALLLIVRR209 pKa = 11.84GMNLGIPYY217 pKa = 10.04ISPALKK223 pKa = 9.74EE224 pKa = 4.32KK225 pKa = 10.61KK226 pKa = 10.2VSCCHH231 pKa = 6.42KK232 pKa = 10.7
MM1 pKa = 7.56NIITSLTYY9 pKa = 10.76LLPGLLLGFLGSFHH23 pKa = 6.9CVGMCGPIALTLPVIHH39 pKa = 6.91LSSTRR44 pKa = 11.84KK45 pKa = 9.34LAGILLYY52 pKa = 10.72NAGRR56 pKa = 11.84TTTYY60 pKa = 11.13LLLGSLSGWLGQRR73 pKa = 11.84FFIGGLQQWLSILLGILLLLLLCWQYY99 pKa = 11.62ILQKK103 pKa = 9.68PLPQIPFFQQHH114 pKa = 5.07VKK116 pKa = 10.4SALGRR121 pKa = 11.84LLTRR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84FSTLFTIGFLNGLLPCGLVYY147 pKa = 10.62IGITGAIATGSAWKK161 pKa = 10.01GALFMTAFGIGTIPAMAAVSWFSQLLRR188 pKa = 11.84VHH190 pKa = 6.66IRR192 pKa = 11.84RR193 pKa = 11.84FIPVAIGMVALLLIVRR209 pKa = 11.84GMNLGIPYY217 pKa = 10.04ISPALKK223 pKa = 9.74EE224 pKa = 4.32KK225 pKa = 10.61KK226 pKa = 10.2VSCCHH231 pKa = 6.42KK232 pKa = 10.7
Molecular weight: 25.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2227784 |
25 |
13566 |
391.6 |
43.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.262 ± 0.043 | 0.86 ± 0.01 |
5.206 ± 0.023 | 5.312 ± 0.037 |
4.355 ± 0.027 | 7.019 ± 0.034 |
2.167 ± 0.02 | 6.695 ± 0.024 |
5.509 ± 0.041 | 9.875 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.27 ± 0.019 | 4.826 ± 0.041 |
4.326 ± 0.027 | 4.552 ± 0.024 |
4.823 ± 0.023 | 5.97 ± 0.025 |
6.061 ± 0.043 | 6.48 ± 0.03 |
1.283 ± 0.012 | 4.149 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
