
Flavobacterium enshiense DK69
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; Flavobacterium enshiense
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
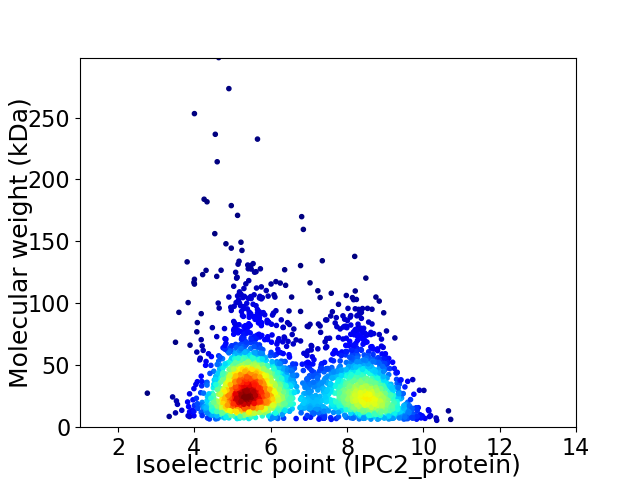
Virtual 2D-PAGE plot for 2848 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V6S848|V6S848_9FLAO Glyoxalase OS=Flavobacterium enshiense DK69 OX=1107311 GN=Q767_13730 PE=4 SV=1
MM1 pKa = 7.8RR2 pKa = 11.84SKK4 pKa = 11.17LLLFYY9 pKa = 10.91GVLFLLFSGNLFGQDD24 pKa = 2.68IALSEE29 pKa = 4.27QFFGRR34 pKa = 11.84YY35 pKa = 9.91DD36 pKa = 3.26FTFFGNTLNPDD47 pKa = 4.03EE48 pKa = 4.48NTFMYY53 pKa = 9.72PDD55 pKa = 4.95SILTSSSADD64 pKa = 3.43LNLSSGSEE72 pKa = 3.73IEE74 pKa = 4.16KK75 pKa = 10.61AYY77 pKa = 10.58LYY79 pKa = 9.65WAGCGPGDD87 pKa = 5.32FNIKK91 pKa = 10.14LNNIDD96 pKa = 3.05IAAQRR101 pKa = 11.84TFSTIQASSGKK112 pKa = 9.69IFFAAFYY119 pKa = 9.8DD120 pKa = 3.34ITDD123 pKa = 3.52IVISQGNTTYY133 pKa = 10.2TVSEE137 pKa = 4.73LDD139 pKa = 3.76VNDD142 pKa = 4.66QIPEE146 pKa = 3.86YY147 pKa = 9.05WDD149 pKa = 3.53NGTHH153 pKa = 5.55FAGWAVLVVYY163 pKa = 10.58KK164 pKa = 11.04NNNLPLNVVNVYY176 pKa = 10.81DD177 pKa = 4.06GLEE180 pKa = 4.17SLSTTPSGGVGEE192 pKa = 4.5LNITIDD198 pKa = 3.92NLNVIDD204 pKa = 5.44DD205 pKa = 4.12VGSKK209 pKa = 9.89IGFIAWEE216 pKa = 4.25GDD218 pKa = 3.04RR219 pKa = 11.84GIPAPGTDD227 pKa = 3.62IIEE230 pKa = 4.3SLSINGITVSNPPLNPANNAFNGTNSVTGAQNLFNMDD267 pKa = 3.56LDD269 pKa = 3.91IYY271 pKa = 10.58NIQNFIDD278 pKa = 4.12IGDD281 pKa = 3.81EE282 pKa = 3.97SATIQLATGQDD293 pKa = 3.37YY294 pKa = 11.87VMINTIVTKK303 pKa = 10.73LNSQLPDD310 pKa = 3.05ATVTIDD316 pKa = 3.77QAEE319 pKa = 4.7GEE321 pKa = 4.56CNSRR325 pKa = 11.84NIAVNYY331 pKa = 9.76SVSNINATDD340 pKa = 4.15FLPAGTPISIYY351 pKa = 10.8AGGILIGTNQTQNDD365 pKa = 3.59IQIGDD370 pKa = 3.96SEE372 pKa = 4.38NFYY375 pKa = 11.22QFVTIPNGVPVNFSLEE391 pKa = 4.25IIVDD395 pKa = 3.6QPGVVTEE402 pKa = 4.16LQEE405 pKa = 4.36NNNNTTMDD413 pKa = 3.49VTLATGPTPNLLADD427 pKa = 5.63LISCNRR433 pKa = 11.84GYY435 pKa = 9.47TEE437 pKa = 4.17GLFDD441 pKa = 4.68FSAYY445 pKa = 10.02EE446 pKa = 3.93NTVTSDD452 pKa = 3.45PNSTVTFHH460 pKa = 7.46EE461 pKa = 4.5SQLEE465 pKa = 4.1ANAGINPIPDD475 pKa = 3.82PSNYY479 pKa = 9.96LATATPKK486 pKa = 10.25TIFVRR491 pKa = 11.84IDD493 pKa = 3.21GPEE496 pKa = 4.09CFTTTSFQLMVKK508 pKa = 8.71PCPPIVYY515 pKa = 9.93NAVSVNGDD523 pKa = 3.19GMNDD527 pKa = 3.34TFHH530 pKa = 8.08IEE532 pKa = 3.75GLRR535 pKa = 11.84DD536 pKa = 3.21VFVNHH541 pKa = 7.26EE542 pKa = 3.74IFIYY546 pKa = 10.18NRR548 pKa = 11.84WGRR551 pKa = 11.84EE552 pKa = 3.54VWKK555 pKa = 10.66GNNYY559 pKa = 7.89TPEE562 pKa = 3.58WDD564 pKa = 4.21GYY566 pKa = 10.16IKK568 pKa = 10.9DD569 pKa = 4.09GVGSNHH575 pKa = 6.56APDD578 pKa = 3.4GTYY581 pKa = 10.27FYY583 pKa = 10.69ILYY586 pKa = 10.33LNDD589 pKa = 3.66PNYY592 pKa = 9.49PKK594 pKa = 10.46PLNGYY599 pKa = 10.01LYY601 pKa = 10.62LNHH604 pKa = 7.19
MM1 pKa = 7.8RR2 pKa = 11.84SKK4 pKa = 11.17LLLFYY9 pKa = 10.91GVLFLLFSGNLFGQDD24 pKa = 2.68IALSEE29 pKa = 4.27QFFGRR34 pKa = 11.84YY35 pKa = 9.91DD36 pKa = 3.26FTFFGNTLNPDD47 pKa = 4.03EE48 pKa = 4.48NTFMYY53 pKa = 9.72PDD55 pKa = 4.95SILTSSSADD64 pKa = 3.43LNLSSGSEE72 pKa = 3.73IEE74 pKa = 4.16KK75 pKa = 10.61AYY77 pKa = 10.58LYY79 pKa = 9.65WAGCGPGDD87 pKa = 5.32FNIKK91 pKa = 10.14LNNIDD96 pKa = 3.05IAAQRR101 pKa = 11.84TFSTIQASSGKK112 pKa = 9.69IFFAAFYY119 pKa = 9.8DD120 pKa = 3.34ITDD123 pKa = 3.52IVISQGNTTYY133 pKa = 10.2TVSEE137 pKa = 4.73LDD139 pKa = 3.76VNDD142 pKa = 4.66QIPEE146 pKa = 3.86YY147 pKa = 9.05WDD149 pKa = 3.53NGTHH153 pKa = 5.55FAGWAVLVVYY163 pKa = 10.58KK164 pKa = 11.04NNNLPLNVVNVYY176 pKa = 10.81DD177 pKa = 4.06GLEE180 pKa = 4.17SLSTTPSGGVGEE192 pKa = 4.5LNITIDD198 pKa = 3.92NLNVIDD204 pKa = 5.44DD205 pKa = 4.12VGSKK209 pKa = 9.89IGFIAWEE216 pKa = 4.25GDD218 pKa = 3.04RR219 pKa = 11.84GIPAPGTDD227 pKa = 3.62IIEE230 pKa = 4.3SLSINGITVSNPPLNPANNAFNGTNSVTGAQNLFNMDD267 pKa = 3.56LDD269 pKa = 3.91IYY271 pKa = 10.58NIQNFIDD278 pKa = 4.12IGDD281 pKa = 3.81EE282 pKa = 3.97SATIQLATGQDD293 pKa = 3.37YY294 pKa = 11.87VMINTIVTKK303 pKa = 10.73LNSQLPDD310 pKa = 3.05ATVTIDD316 pKa = 3.77QAEE319 pKa = 4.7GEE321 pKa = 4.56CNSRR325 pKa = 11.84NIAVNYY331 pKa = 9.76SVSNINATDD340 pKa = 4.15FLPAGTPISIYY351 pKa = 10.8AGGILIGTNQTQNDD365 pKa = 3.59IQIGDD370 pKa = 3.96SEE372 pKa = 4.38NFYY375 pKa = 11.22QFVTIPNGVPVNFSLEE391 pKa = 4.25IIVDD395 pKa = 3.6QPGVVTEE402 pKa = 4.16LQEE405 pKa = 4.36NNNNTTMDD413 pKa = 3.49VTLATGPTPNLLADD427 pKa = 5.63LISCNRR433 pKa = 11.84GYY435 pKa = 9.47TEE437 pKa = 4.17GLFDD441 pKa = 4.68FSAYY445 pKa = 10.02EE446 pKa = 3.93NTVTSDD452 pKa = 3.45PNSTVTFHH460 pKa = 7.46EE461 pKa = 4.5SQLEE465 pKa = 4.1ANAGINPIPDD475 pKa = 3.82PSNYY479 pKa = 9.96LATATPKK486 pKa = 10.25TIFVRR491 pKa = 11.84IDD493 pKa = 3.21GPEE496 pKa = 4.09CFTTTSFQLMVKK508 pKa = 8.71PCPPIVYY515 pKa = 9.93NAVSVNGDD523 pKa = 3.19GMNDD527 pKa = 3.34TFHH530 pKa = 8.08IEE532 pKa = 3.75GLRR535 pKa = 11.84DD536 pKa = 3.21VFVNHH541 pKa = 7.26EE542 pKa = 3.74IFIYY546 pKa = 10.18NRR548 pKa = 11.84WGRR551 pKa = 11.84EE552 pKa = 3.54VWKK555 pKa = 10.66GNNYY559 pKa = 7.89TPEE562 pKa = 3.58WDD564 pKa = 4.21GYY566 pKa = 10.16IKK568 pKa = 10.9DD569 pKa = 4.09GVGSNHH575 pKa = 6.56APDD578 pKa = 3.4GTYY581 pKa = 10.27FYY583 pKa = 10.69ILYY586 pKa = 10.33LNDD589 pKa = 3.66PNYY592 pKa = 9.49PKK594 pKa = 10.46PLNGYY599 pKa = 10.01LYY601 pKa = 10.62LNHH604 pKa = 7.19
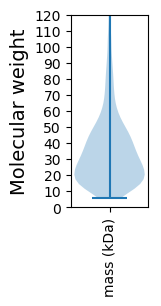
Molecular weight: 66.23 kDa
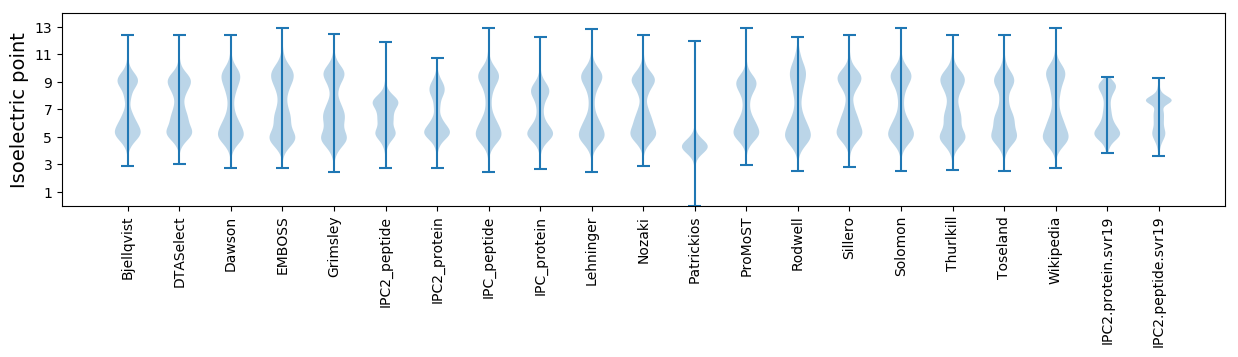
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V6S8U0|V6S8U0_9FLAO Uncharacterized protein OS=Flavobacterium enshiense DK69 OX=1107311 GN=Q767_07315 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
932535 |
46 |
2947 |
327.4 |
36.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.527 ± 0.06 | 0.836 ± 0.016 |
5.307 ± 0.035 | 6.499 ± 0.06 |
5.361 ± 0.044 | 6.518 ± 0.057 |
1.744 ± 0.021 | 7.756 ± 0.051 |
7.871 ± 0.074 | 8.968 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.358 ± 0.025 | 6.149 ± 0.049 |
3.5 ± 0.03 | 3.349 ± 0.027 |
3.242 ± 0.029 | 6.463 ± 0.043 |
6.012 ± 0.086 | 6.516 ± 0.039 |
1.005 ± 0.017 | 4.02 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
