
Porphyromonas sp. COT-239 OH1446
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Porphyromonadaceae; Porphyromonas; unclassified Porphyromonas
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
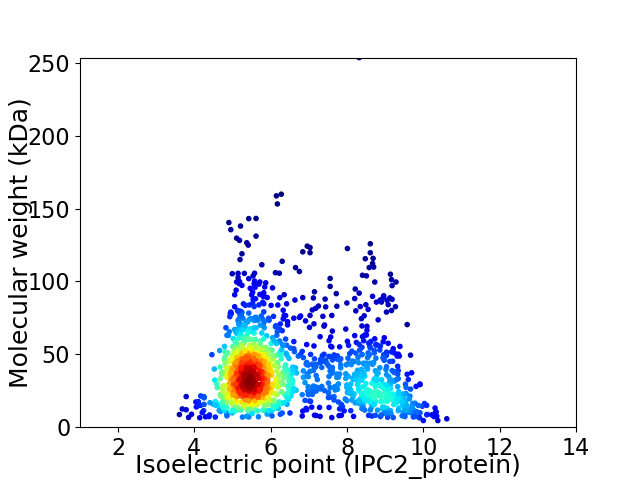
Virtual 2D-PAGE plot for 1412 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A2E0K7|A0A0A2E0K7_9PORP Uncharacterized protein OS=Porphyromonas sp. COT-239 OH1446 OX=1515613 GN=HQ37_03290 PE=4 SV=1
MM1 pKa = 7.85PEE3 pKa = 3.37VALQFVEE10 pKa = 5.05EE11 pKa = 4.09VLPYY15 pKa = 10.8GDD17 pKa = 3.35VFAFEE22 pKa = 5.24APMGTGKK29 pKa = 7.7TTFIKK34 pKa = 10.52ALCLALGVQDD44 pKa = 4.85VINSPTFSIINEE56 pKa = 3.82YY57 pKa = 9.92RR58 pKa = 11.84AEE60 pKa = 4.1PSGEE64 pKa = 4.16LIYY67 pKa = 11.12HH68 pKa = 6.23FDD70 pKa = 3.62CYY72 pKa = 10.64RR73 pKa = 11.84LEE75 pKa = 4.32RR76 pKa = 11.84LDD78 pKa = 4.51DD79 pKa = 4.31ALSLGVEE86 pKa = 4.89DD87 pKa = 4.38YY88 pKa = 11.37LEE90 pKa = 4.54SGALCLIEE98 pKa = 4.15WPKK101 pKa = 10.8LLEE104 pKa = 5.11PILPSDD110 pKa = 3.71TVLVEE115 pKa = 4.62LSLEE119 pKa = 4.05ADD121 pKa = 3.53GQGRR125 pKa = 11.84ILTAHH130 pKa = 5.8YY131 pKa = 11.15NEE133 pKa = 4.47VV134 pKa = 3.15
MM1 pKa = 7.85PEE3 pKa = 3.37VALQFVEE10 pKa = 5.05EE11 pKa = 4.09VLPYY15 pKa = 10.8GDD17 pKa = 3.35VFAFEE22 pKa = 5.24APMGTGKK29 pKa = 7.7TTFIKK34 pKa = 10.52ALCLALGVQDD44 pKa = 4.85VINSPTFSIINEE56 pKa = 3.82YY57 pKa = 9.92RR58 pKa = 11.84AEE60 pKa = 4.1PSGEE64 pKa = 4.16LIYY67 pKa = 11.12HH68 pKa = 6.23FDD70 pKa = 3.62CYY72 pKa = 10.64RR73 pKa = 11.84LEE75 pKa = 4.32RR76 pKa = 11.84LDD78 pKa = 4.51DD79 pKa = 4.31ALSLGVEE86 pKa = 4.89DD87 pKa = 4.38YY88 pKa = 11.37LEE90 pKa = 4.54SGALCLIEE98 pKa = 4.15WPKK101 pKa = 10.8LLEE104 pKa = 5.11PILPSDD110 pKa = 3.71TVLVEE115 pKa = 4.62LSLEE119 pKa = 4.05ADD121 pKa = 3.53GQGRR125 pKa = 11.84ILTAHH130 pKa = 5.8YY131 pKa = 11.15NEE133 pKa = 4.47VV134 pKa = 3.15
Molecular weight: 14.88 kDa
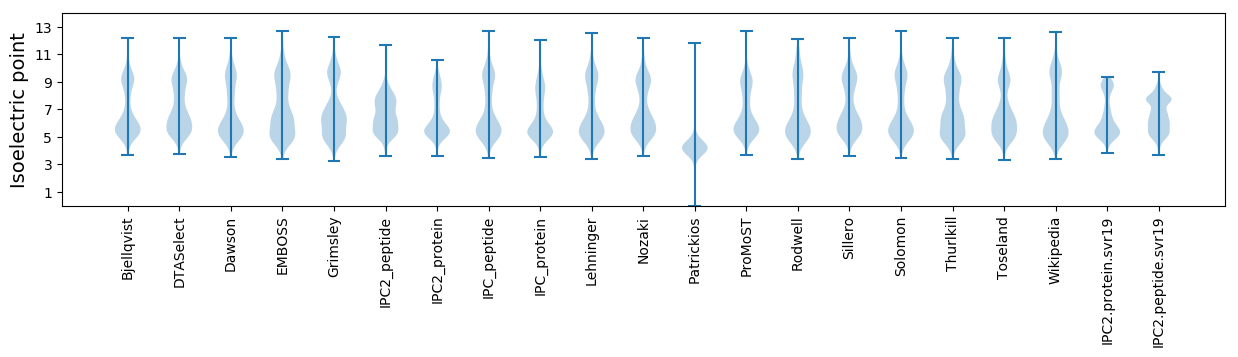
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A2DX28|A0A0A2DX28_9PORP Beta-N-acetylhexosaminidase OS=Porphyromonas sp. COT-239 OH1446 OX=1515613 GN=HQ37_05180 PE=3 SV=1
MM1 pKa = 7.57KK2 pKa = 9.99IARR5 pKa = 11.84VTPLMRR11 pKa = 11.84ALLLFIACLCAVNSLSAQGFSLSSGSSPQPRR42 pKa = 11.84PTDD45 pKa = 3.15SLRR48 pKa = 11.84ISRR51 pKa = 11.84RR52 pKa = 11.84LRR54 pKa = 11.84PEE56 pKa = 3.64VKK58 pKa = 10.15SRR60 pKa = 11.84LLPQQSYY67 pKa = 10.73GINAHH72 pKa = 6.22RR73 pKa = 11.84RR74 pKa = 11.84TMLIRR79 pKa = 11.84PEE81 pKa = 4.2FGVLPIRR88 pKa = 11.84DD89 pKa = 3.63GSTIILNGEE98 pKa = 4.06TPIASYY104 pKa = 10.93EE105 pKa = 4.03PLDD108 pKa = 4.14PRR110 pKa = 11.84PSSIRR115 pKa = 11.84DD116 pKa = 3.24LSPRR120 pKa = 11.84GIRR123 pKa = 11.84QSSGIRR129 pKa = 11.84LMGRR133 pKa = 11.84TLYY136 pKa = 10.76FSDD139 pKa = 3.32PAEE142 pKa = 3.97RR143 pKa = 11.84VEE145 pKa = 5.5LIDD148 pKa = 3.42VTGKK152 pKa = 10.35VVLAYY157 pKa = 10.69RR158 pKa = 11.84NVRR161 pKa = 11.84EE162 pKa = 4.27AKK164 pKa = 10.42LNLPEE169 pKa = 5.11GIYY172 pKa = 10.39IMRR175 pKa = 11.84CNINDD180 pKa = 4.16DD181 pKa = 4.02YY182 pKa = 11.31TVSRR186 pKa = 11.84LILRR190 pKa = 4.21
MM1 pKa = 7.57KK2 pKa = 9.99IARR5 pKa = 11.84VTPLMRR11 pKa = 11.84ALLLFIACLCAVNSLSAQGFSLSSGSSPQPRR42 pKa = 11.84PTDD45 pKa = 3.15SLRR48 pKa = 11.84ISRR51 pKa = 11.84RR52 pKa = 11.84LRR54 pKa = 11.84PEE56 pKa = 3.64VKK58 pKa = 10.15SRR60 pKa = 11.84LLPQQSYY67 pKa = 10.73GINAHH72 pKa = 6.22RR73 pKa = 11.84RR74 pKa = 11.84TMLIRR79 pKa = 11.84PEE81 pKa = 4.2FGVLPIRR88 pKa = 11.84DD89 pKa = 3.63GSTIILNGEE98 pKa = 4.06TPIASYY104 pKa = 10.93EE105 pKa = 4.03PLDD108 pKa = 4.14PRR110 pKa = 11.84PSSIRR115 pKa = 11.84DD116 pKa = 3.24LSPRR120 pKa = 11.84GIRR123 pKa = 11.84QSSGIRR129 pKa = 11.84LMGRR133 pKa = 11.84TLYY136 pKa = 10.76FSDD139 pKa = 3.32PAEE142 pKa = 3.97RR143 pKa = 11.84VEE145 pKa = 5.5LIDD148 pKa = 3.42VTGKK152 pKa = 10.35VVLAYY157 pKa = 10.69RR158 pKa = 11.84NVRR161 pKa = 11.84EE162 pKa = 4.27AKK164 pKa = 10.42LNLPEE169 pKa = 5.11GIYY172 pKa = 10.39IMRR175 pKa = 11.84CNINDD180 pKa = 4.16DD181 pKa = 4.02YY182 pKa = 11.31TVSRR186 pKa = 11.84LILRR190 pKa = 4.21
Molecular weight: 21.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
513336 |
38 |
2236 |
363.6 |
40.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.121 ± 0.069 | 1.01 ± 0.024 |
5.003 ± 0.041 | 7.186 ± 0.066 |
3.774 ± 0.035 | 7.315 ± 0.047 |
2.311 ± 0.025 | 6.32 ± 0.048 |
4.533 ± 0.064 | 10.975 ± 0.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.544 ± 0.031 | 3.242 ± 0.043 |
4.233 ± 0.038 | 3.837 ± 0.036 |
6.716 ± 0.053 | 6.851 ± 0.063 |
4.744 ± 0.033 | 6.207 ± 0.046 |
1.131 ± 0.025 | 3.947 ± 0.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
