
MW polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Deltapolyomavirus; Human polyomavirus 10
Average proteome isoelectric point is 5.5
Get precalculated fractions of proteins
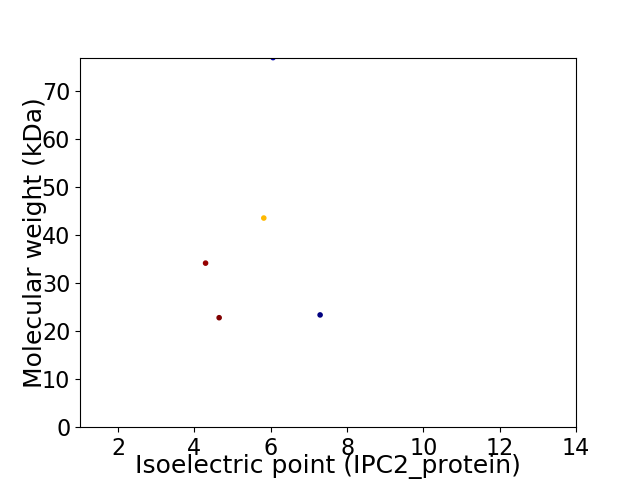
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
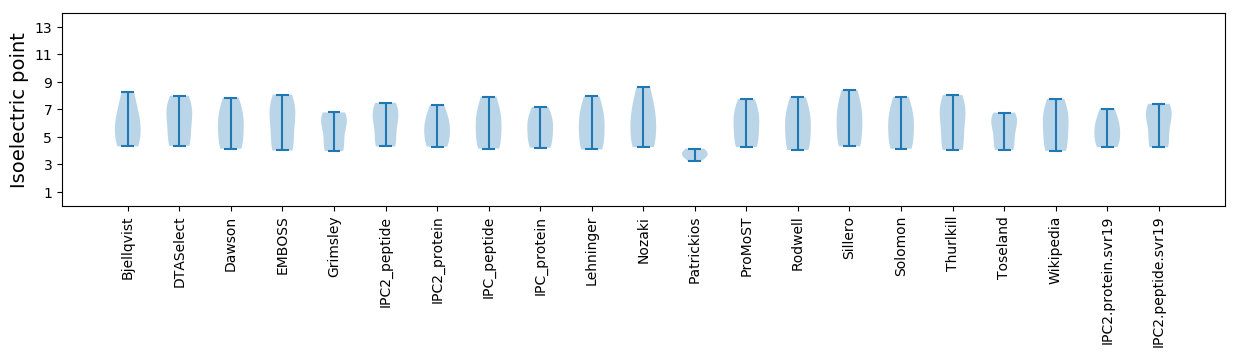
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I6UY22|I6UY22_9POLY Large T antigen OS=MW polyomavirus OX=1203539 PE=4 SV=1
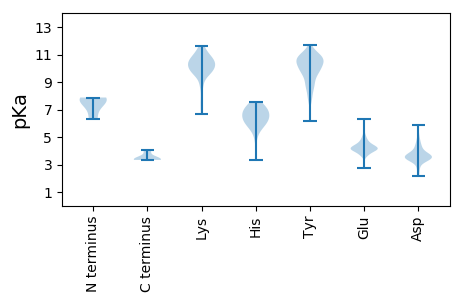
MM1 pKa = 7.29GAVISVILDD10 pKa = 4.3LIEE13 pKa = 5.14LISVTGFEE21 pKa = 4.71AEE23 pKa = 4.31TIISGEE29 pKa = 3.54AAAIVEE35 pKa = 4.52SQLSSLAVIEE45 pKa = 4.31NASAAEE51 pKa = 3.99VLSTFGLNEE60 pKa = 3.75TSYY63 pKa = 11.66SLLVNFPQAFEE74 pKa = 4.13NAVYY78 pKa = 8.66TAQLIQTISGASSLIAAGIQTQPFQIFDD106 pKa = 3.71AGSNMALQQWRR117 pKa = 11.84PDD119 pKa = 3.58YY120 pKa = 10.97FDD122 pKa = 3.94VFIPGYY128 pKa = 9.98RR129 pKa = 11.84HH130 pKa = 5.73FEE132 pKa = 4.15YY133 pKa = 10.88YY134 pKa = 10.85FNVLSGWGEE143 pKa = 4.1SLVNTVSRR151 pKa = 11.84AFWEE155 pKa = 4.22ALLSEE160 pKa = 4.52TRR162 pKa = 11.84QTARR166 pKa = 11.84LLASNAVDD174 pKa = 3.06NVYY177 pKa = 11.03NVGEE181 pKa = 4.09QGLQNIQNALVGLIEE196 pKa = 4.09GARR199 pKa = 11.84WALRR203 pKa = 11.84FPGNVYY209 pKa = 10.63HH210 pKa = 6.52NLEE213 pKa = 4.07MYY215 pKa = 9.79YY216 pKa = 10.78AQLPGLTPPQVRR228 pKa = 11.84SIQKK232 pKa = 9.8RR233 pKa = 11.84LEE235 pKa = 3.7EE236 pKa = 4.01ARR238 pKa = 11.84NYY240 pKa = 9.78GPSVQVDD247 pKa = 3.45DD248 pKa = 4.83SEE250 pKa = 4.88NSVFSEE256 pKa = 4.46HH257 pKa = 7.16LSGDD261 pKa = 3.66YY262 pKa = 10.34IFRR265 pKa = 11.84GEE267 pKa = 4.13APGGARR273 pKa = 11.84QRR275 pKa = 11.84QTPDD279 pKa = 2.15WMLQLILGILGDD291 pKa = 3.39ITPFFKK297 pKa = 10.61EE298 pKa = 3.79VIEE301 pKa = 4.06EE302 pKa = 4.32VEE304 pKa = 4.14GEE306 pKa = 4.26EE307 pKa = 4.95NADD310 pKa = 3.36
MM1 pKa = 7.29GAVISVILDD10 pKa = 4.3LIEE13 pKa = 5.14LISVTGFEE21 pKa = 4.71AEE23 pKa = 4.31TIISGEE29 pKa = 3.54AAAIVEE35 pKa = 4.52SQLSSLAVIEE45 pKa = 4.31NASAAEE51 pKa = 3.99VLSTFGLNEE60 pKa = 3.75TSYY63 pKa = 11.66SLLVNFPQAFEE74 pKa = 4.13NAVYY78 pKa = 8.66TAQLIQTISGASSLIAAGIQTQPFQIFDD106 pKa = 3.71AGSNMALQQWRR117 pKa = 11.84PDD119 pKa = 3.58YY120 pKa = 10.97FDD122 pKa = 3.94VFIPGYY128 pKa = 9.98RR129 pKa = 11.84HH130 pKa = 5.73FEE132 pKa = 4.15YY133 pKa = 10.88YY134 pKa = 10.85FNVLSGWGEE143 pKa = 4.1SLVNTVSRR151 pKa = 11.84AFWEE155 pKa = 4.22ALLSEE160 pKa = 4.52TRR162 pKa = 11.84QTARR166 pKa = 11.84LLASNAVDD174 pKa = 3.06NVYY177 pKa = 11.03NVGEE181 pKa = 4.09QGLQNIQNALVGLIEE196 pKa = 4.09GARR199 pKa = 11.84WALRR203 pKa = 11.84FPGNVYY209 pKa = 10.63HH210 pKa = 6.52NLEE213 pKa = 4.07MYY215 pKa = 9.79YY216 pKa = 10.78AQLPGLTPPQVRR228 pKa = 11.84SIQKK232 pKa = 9.8RR233 pKa = 11.84LEE235 pKa = 3.7EE236 pKa = 4.01ARR238 pKa = 11.84NYY240 pKa = 9.78GPSVQVDD247 pKa = 3.45DD248 pKa = 4.83SEE250 pKa = 4.88NSVFSEE256 pKa = 4.46HH257 pKa = 7.16LSGDD261 pKa = 3.66YY262 pKa = 10.34IFRR265 pKa = 11.84GEE267 pKa = 4.13APGGARR273 pKa = 11.84QRR275 pKa = 11.84QTPDD279 pKa = 2.15WMLQLILGILGDD291 pKa = 3.39ITPFFKK297 pKa = 10.61EE298 pKa = 3.79VIEE301 pKa = 4.06EE302 pKa = 4.32VEE304 pKa = 4.14GEE306 pKa = 4.26EE307 pKa = 4.95NADD310 pKa = 3.36
Molecular weight: 34.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I6ULL7|I6ULL7_9POLY Minor capsid protein VP2 OS=MW polyomavirus OX=1203539 GN=VP3 PE=3 SV=1
MM1 pKa = 7.79DD2 pKa = 4.63RR3 pKa = 11.84VLSRR7 pKa = 11.84DD8 pKa = 3.33EE9 pKa = 4.46VKK11 pKa = 10.96EE12 pKa = 3.79LMALLSLNVAAWGNIPLMQYY32 pKa = 10.52KK33 pKa = 9.65YY34 pKa = 10.65RR35 pKa = 11.84QACLKK40 pKa = 9.6LHH42 pKa = 6.86PDD44 pKa = 3.24KK45 pKa = 11.65GGDD48 pKa = 3.6GEE50 pKa = 4.46KK51 pKa = 9.62MKK53 pKa = 10.69RR54 pKa = 11.84LNEE57 pKa = 4.33LFSKK61 pKa = 10.09MYY63 pKa = 7.8TTIEE67 pKa = 3.79KK68 pKa = 10.13LRR70 pKa = 11.84RR71 pKa = 11.84EE72 pKa = 4.34GEE74 pKa = 4.26VYY76 pKa = 10.34FPAKK80 pKa = 9.68VGYY83 pKa = 9.81FIEE86 pKa = 4.88DD87 pKa = 3.71VVTLGDD93 pKa = 3.53VLGPTFEE100 pKa = 4.36EE101 pKa = 4.37KK102 pKa = 9.93IIYY105 pKa = 9.13IWPLCASDD113 pKa = 5.1LVRR116 pKa = 11.84HH117 pKa = 5.96KK118 pKa = 10.45CGCVCCLLKK127 pKa = 10.55KK128 pKa = 7.61QHH130 pKa = 6.77RR131 pKa = 11.84NDD133 pKa = 3.35KK134 pKa = 10.17LAKK137 pKa = 9.6QKK139 pKa = 10.56QCLVWGEE146 pKa = 4.31CFCYY150 pKa = 10.55SCFLLWFGQEE160 pKa = 3.73FGYY163 pKa = 9.63TSFFWWKK170 pKa = 10.33HH171 pKa = 3.34IMHH174 pKa = 7.09NIEE177 pKa = 4.23FDD179 pKa = 3.79LLRR182 pKa = 11.84LLGEE186 pKa = 4.68LILWVSVSFFILGG199 pKa = 3.34
MM1 pKa = 7.79DD2 pKa = 4.63RR3 pKa = 11.84VLSRR7 pKa = 11.84DD8 pKa = 3.33EE9 pKa = 4.46VKK11 pKa = 10.96EE12 pKa = 3.79LMALLSLNVAAWGNIPLMQYY32 pKa = 10.52KK33 pKa = 9.65YY34 pKa = 10.65RR35 pKa = 11.84QACLKK40 pKa = 9.6LHH42 pKa = 6.86PDD44 pKa = 3.24KK45 pKa = 11.65GGDD48 pKa = 3.6GEE50 pKa = 4.46KK51 pKa = 9.62MKK53 pKa = 10.69RR54 pKa = 11.84LNEE57 pKa = 4.33LFSKK61 pKa = 10.09MYY63 pKa = 7.8TTIEE67 pKa = 3.79KK68 pKa = 10.13LRR70 pKa = 11.84RR71 pKa = 11.84EE72 pKa = 4.34GEE74 pKa = 4.26VYY76 pKa = 10.34FPAKK80 pKa = 9.68VGYY83 pKa = 9.81FIEE86 pKa = 4.88DD87 pKa = 3.71VVTLGDD93 pKa = 3.53VLGPTFEE100 pKa = 4.36EE101 pKa = 4.37KK102 pKa = 9.93IIYY105 pKa = 9.13IWPLCASDD113 pKa = 5.1LVRR116 pKa = 11.84HH117 pKa = 5.96KK118 pKa = 10.45CGCVCCLLKK127 pKa = 10.55KK128 pKa = 7.61QHH130 pKa = 6.77RR131 pKa = 11.84NDD133 pKa = 3.35KK134 pKa = 10.17LAKK137 pKa = 9.6QKK139 pKa = 10.56QCLVWGEE146 pKa = 4.31CFCYY150 pKa = 10.55SCFLLWFGQEE160 pKa = 3.73FGYY163 pKa = 9.63TSFFWWKK170 pKa = 10.33HH171 pKa = 3.34IMHH174 pKa = 7.09NIEE177 pKa = 4.23FDD179 pKa = 3.79LLRR182 pKa = 11.84LLGEE186 pKa = 4.68LILWVSVSFFILGG199 pKa = 3.34
Molecular weight: 23.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1780 |
199 |
668 |
356.0 |
40.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.73 ± 0.901 | 2.697 ± 0.706 |
5.393 ± 0.556 | 6.798 ± 1.034 |
4.494 ± 0.679 | 6.629 ± 0.64 |
1.629 ± 0.253 | 5.112 ± 0.48 |
5.449 ± 1.472 | 10.169 ± 0.846 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.36 ± 0.274 | 4.944 ± 0.448 |
5.73 ± 1.171 | 4.494 ± 0.512 |
4.607 ± 0.292 | 5.787 ± 0.524 |
5.562 ± 1.324 | 7.022 ± 0.723 |
1.742 ± 0.331 | 3.652 ± 0.238 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
