
Spodoptera exigua multiple nucleopolyhedrovirus
Taxonomy: Viruses; Naldaviricetes; Lefavirales; Baculoviridae; Alphabaculovirus
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
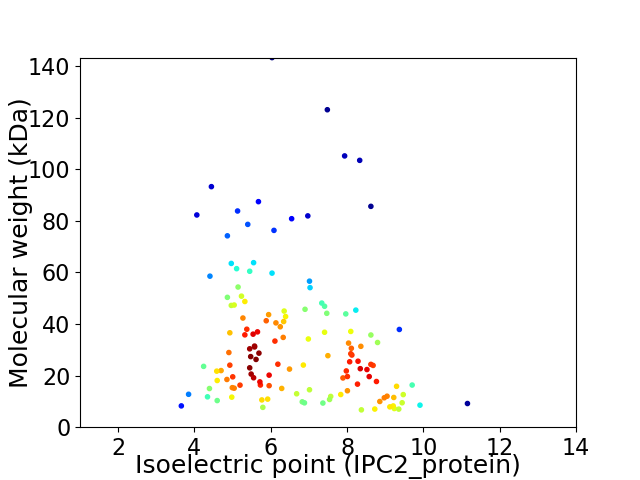
Virtual 2D-PAGE plot for 135 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9J833|Q9J833_9ABAC ORF104 OS=Spodoptera exigua multiple nucleopolyhedrovirus OX=10454 GN=HTSEG24_101 PE=4 SV=1
MM1 pKa = 7.4SSVSIANDD9 pKa = 3.04VLNAILSDD17 pKa = 3.75NLEE20 pKa = 5.13LIDD23 pKa = 5.22DD24 pKa = 4.31SYY26 pKa = 11.78IILNVVDD33 pKa = 4.76HH34 pKa = 6.81EE35 pKa = 4.9SSGGTIKK42 pKa = 10.21PVCIGEE48 pKa = 4.17INSFQTNQSDD58 pKa = 4.15KK59 pKa = 10.98YY60 pKa = 8.88PVSDD64 pKa = 3.58SSVSSEE70 pKa = 4.05LQSDD74 pKa = 3.58QTLL77 pKa = 3.3
MM1 pKa = 7.4SSVSIANDD9 pKa = 3.04VLNAILSDD17 pKa = 3.75NLEE20 pKa = 5.13LIDD23 pKa = 5.22DD24 pKa = 4.31SYY26 pKa = 11.78IILNVVDD33 pKa = 4.76HH34 pKa = 6.81EE35 pKa = 4.9SSGGTIKK42 pKa = 10.21PVCIGEE48 pKa = 4.17INSFQTNQSDD58 pKa = 4.15KK59 pKa = 10.98YY60 pKa = 8.88PVSDD64 pKa = 3.58SSVSSEE70 pKa = 4.05LQSDD74 pKa = 3.58QTLL77 pKa = 3.3
Molecular weight: 8.26 kDa
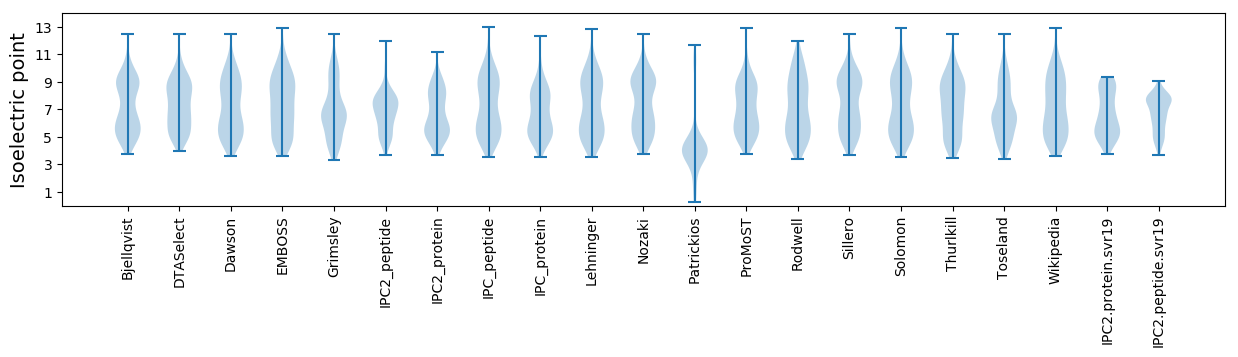
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9J891|Q9J891_9ABAC ORF44 OS=Spodoptera exigua multiple nucleopolyhedrovirus OX=10454 PE=4 SV=1
MM1 pKa = 7.43WLISKK6 pKa = 9.66KK7 pKa = 10.49CQDD10 pKa = 3.03INVSVFFDD18 pKa = 3.43QFCVMWVSADD28 pKa = 3.77DD29 pKa = 3.83VLHH32 pKa = 6.85LLRR35 pKa = 11.84LPASTLQNIPQRR47 pKa = 11.84HH48 pKa = 5.15KK49 pKa = 10.79KK50 pKa = 9.17CWVDD54 pKa = 4.55FRR56 pKa = 11.84CPHH59 pKa = 6.33HH60 pKa = 6.92CSHH63 pKa = 7.74DD64 pKa = 3.63GGKK67 pKa = 9.99VFIDD71 pKa = 4.44LYY73 pKa = 11.61GLGNLCNRR81 pKa = 11.84VNSQIADD88 pKa = 3.48YY89 pKa = 11.33LMTVFVAEE97 pKa = 4.49SYY99 pKa = 10.69NEE101 pKa = 3.59RR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84DD105 pKa = 3.35SRR107 pKa = 11.84RR108 pKa = 11.84SFSPRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84SHH117 pKa = 4.85SPRR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84SPSPRR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84SPSPRR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84SRR138 pKa = 11.84SRR140 pKa = 11.84SGGGGAGRR148 pKa = 11.84RR149 pKa = 11.84RR150 pKa = 11.84SSSRR154 pKa = 11.84CCPRR158 pKa = 11.84RR159 pKa = 11.84HH160 pKa = 5.82HH161 pKa = 6.35HH162 pKa = 6.13HH163 pKa = 6.5HH164 pKa = 6.37HH165 pKa = 6.01HH166 pKa = 6.37HH167 pKa = 5.66HH168 pKa = 6.44TEE170 pKa = 3.54LLEE173 pKa = 4.81RR174 pKa = 11.84ITRR177 pKa = 11.84QNDD180 pKa = 4.15LILTSVNQLTVTNANQHH197 pKa = 6.26LEE199 pKa = 3.92LSNILNALRR208 pKa = 11.84LQQTTIAGQVAQILEE223 pKa = 4.42TVEE226 pKa = 3.97GLGDD230 pKa = 3.28ITGDD234 pKa = 3.48FTRR237 pKa = 11.84LLAEE241 pKa = 4.63LDD243 pKa = 3.43TRR245 pKa = 11.84LAALSTTILNAINQLSDD262 pKa = 3.16QLRR265 pKa = 11.84NEE267 pKa = 3.94LTGINSVLNNLSSSVTNINATLNNLLQAINGLNLGTVVGEE307 pKa = 4.05LQQTINTILDD317 pKa = 4.09LLQTILGILQPNIPLNKK334 pKa = 9.73KK335 pKa = 9.61
MM1 pKa = 7.43WLISKK6 pKa = 9.66KK7 pKa = 10.49CQDD10 pKa = 3.03INVSVFFDD18 pKa = 3.43QFCVMWVSADD28 pKa = 3.77DD29 pKa = 3.83VLHH32 pKa = 6.85LLRR35 pKa = 11.84LPASTLQNIPQRR47 pKa = 11.84HH48 pKa = 5.15KK49 pKa = 10.79KK50 pKa = 9.17CWVDD54 pKa = 4.55FRR56 pKa = 11.84CPHH59 pKa = 6.33HH60 pKa = 6.92CSHH63 pKa = 7.74DD64 pKa = 3.63GGKK67 pKa = 9.99VFIDD71 pKa = 4.44LYY73 pKa = 11.61GLGNLCNRR81 pKa = 11.84VNSQIADD88 pKa = 3.48YY89 pKa = 11.33LMTVFVAEE97 pKa = 4.49SYY99 pKa = 10.69NEE101 pKa = 3.59RR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84DD105 pKa = 3.35SRR107 pKa = 11.84RR108 pKa = 11.84SFSPRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84SHH117 pKa = 4.85SPRR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84SPSPRR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84SPSPRR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84SRR138 pKa = 11.84SRR140 pKa = 11.84SGGGGAGRR148 pKa = 11.84RR149 pKa = 11.84RR150 pKa = 11.84SSSRR154 pKa = 11.84CCPRR158 pKa = 11.84RR159 pKa = 11.84HH160 pKa = 5.82HH161 pKa = 6.35HH162 pKa = 6.13HH163 pKa = 6.5HH164 pKa = 6.37HH165 pKa = 6.01HH166 pKa = 6.37HH167 pKa = 5.66HH168 pKa = 6.44TEE170 pKa = 3.54LLEE173 pKa = 4.81RR174 pKa = 11.84ITRR177 pKa = 11.84QNDD180 pKa = 4.15LILTSVNQLTVTNANQHH197 pKa = 6.26LEE199 pKa = 3.92LSNILNALRR208 pKa = 11.84LQQTTIAGQVAQILEE223 pKa = 4.42TVEE226 pKa = 3.97GLGDD230 pKa = 3.28ITGDD234 pKa = 3.48FTRR237 pKa = 11.84LLAEE241 pKa = 4.63LDD243 pKa = 3.43TRR245 pKa = 11.84LAALSTTILNAINQLSDD262 pKa = 3.16QLRR265 pKa = 11.84NEE267 pKa = 3.94LTGINSVLNNLSSSVTNINATLNNLLQAINGLNLGTVVGEE307 pKa = 4.05LQQTINTILDD317 pKa = 4.09LLQTILGILQPNIPLNKK334 pKa = 9.73KK335 pKa = 9.61
Molecular weight: 37.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
39664 |
57 |
1222 |
293.8 |
33.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.115 ± 0.19 | 2.224 ± 0.129 |
7.319 ± 0.251 | 5.065 ± 0.116 |
4.856 ± 0.141 | 3.477 ± 0.153 |
2.327 ± 0.103 | 6.837 ± 0.133 |
6.593 ± 0.287 | 8.728 ± 0.198 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.753 ± 0.102 | 7.558 ± 0.168 |
3.515 ± 0.188 | 3.457 ± 0.179 |
5.206 ± 0.148 | 6.648 ± 0.199 |
5.695 ± 0.165 | 7.092 ± 0.153 |
0.711 ± 0.054 | 4.823 ± 0.141 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
