
Geodermatophilus ruber
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Geodermatophilus
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
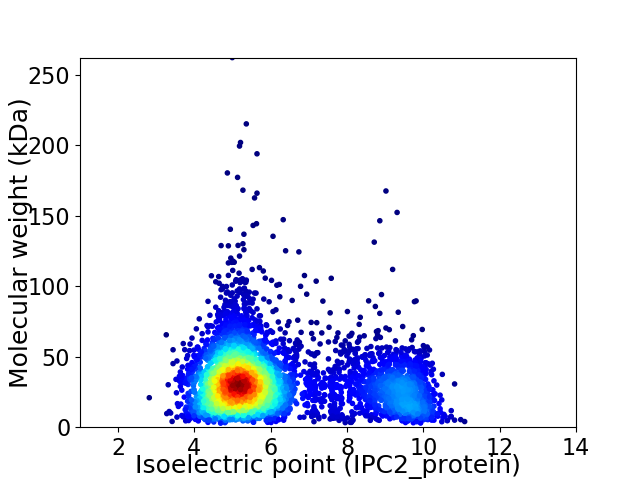
Virtual 2D-PAGE plot for 4702 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4IIM6|A0A1I4IIM6_9ACTN Cob(I)alamin adenosyltransferase OS=Geodermatophilus ruber OX=504800 GN=SAMN04488085_11311 PE=4 SV=1
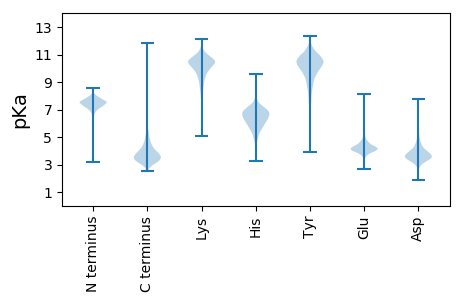
MM1 pKa = 7.35RR2 pKa = 11.84QVQRR6 pKa = 11.84RR7 pKa = 11.84ALAAAAAGSMLLAGCATTVTGTAAAAHH34 pKa = 7.06PLTDD38 pKa = 3.62VPAEE42 pKa = 4.01ALPITGAGDD51 pKa = 3.46GAIDD55 pKa = 3.6VLARR59 pKa = 11.84NAMADD64 pKa = 3.47LLTYY68 pKa = 9.05WEE70 pKa = 3.99RR71 pKa = 11.84TYY73 pKa = 11.11PEE75 pKa = 5.08LYY77 pKa = 10.65GEE79 pKa = 4.31PFVPLEE85 pKa = 4.1GGIFAVDD92 pKa = 4.58ADD94 pKa = 4.33DD95 pKa = 5.91LDD97 pKa = 4.22PAVYY101 pKa = 9.4PDD103 pKa = 3.83TGIGCDD109 pKa = 4.08RR110 pKa = 11.84LPVDD114 pKa = 3.61PSEE117 pKa = 4.46VEE119 pKa = 4.17GNAFYY124 pKa = 11.15LLPCDD129 pKa = 3.96LVAYY133 pKa = 10.05DD134 pKa = 3.62SALIEE139 pKa = 4.21EE140 pKa = 4.84LAAAHH145 pKa = 6.44GRR147 pKa = 11.84FLGPAVMAHH156 pKa = 5.74EE157 pKa = 5.05MGHH160 pKa = 7.21AIQGRR165 pKa = 11.84NGTWDD170 pKa = 3.43TVDD173 pKa = 3.88TIVSEE178 pKa = 4.33TQADD182 pKa = 4.12CFAGAWTRR190 pKa = 11.84WVADD194 pKa = 3.49GNAAHH199 pKa = 6.75SSLRR203 pKa = 11.84VPEE206 pKa = 4.26LDD208 pKa = 3.29EE209 pKa = 4.21VVVGFLEE216 pKa = 4.49LRR218 pKa = 11.84DD219 pKa = 3.84PVGTGTDD226 pKa = 3.02EE227 pKa = 5.08DD228 pKa = 4.62GAHH231 pKa = 6.62GSGFDD236 pKa = 3.23RR237 pKa = 11.84VSGFFAGWDD246 pKa = 3.63GGATACRR253 pKa = 11.84DD254 pKa = 3.5EE255 pKa = 4.53FDD257 pKa = 3.56EE258 pKa = 5.74DD259 pKa = 4.2RR260 pKa = 11.84LFTAAEE266 pKa = 4.17FTDD269 pKa = 4.53HH270 pKa = 7.29GDD272 pKa = 4.33LEE274 pKa = 4.73NEE276 pKa = 4.14GNASYY281 pKa = 11.35DD282 pKa = 3.57EE283 pKa = 4.21TLLIVDD289 pKa = 4.2ASLPLFYY296 pKa = 10.86EE297 pKa = 4.53SIFPSEE303 pKa = 3.97FGTRR307 pKa = 11.84FQVPALEE314 pKa = 4.9AFDD317 pKa = 4.48GTAPDD322 pKa = 4.0CGEE325 pKa = 4.39LGAHH329 pKa = 6.23GRR331 pKa = 11.84DD332 pKa = 3.5LGYY335 pKa = 10.6CGADD339 pKa = 3.0GTVYY343 pKa = 10.55FDD345 pKa = 3.51EE346 pKa = 4.98TDD348 pKa = 4.21LIEE351 pKa = 4.17PAYY354 pKa = 10.7AEE356 pKa = 4.58LGDD359 pKa = 3.78FAVATGIALPYY370 pKa = 10.52ALAARR375 pKa = 11.84DD376 pKa = 3.78QLGLSTDD383 pKa = 3.72DD384 pKa = 3.75GAATRR389 pKa = 11.84SAVCLTGWYY398 pKa = 7.87TADD401 pKa = 3.82FFAGDD406 pKa = 3.71FRR408 pKa = 11.84EE409 pKa = 4.5VTSLSPGDD417 pKa = 3.31VDD419 pKa = 3.7EE420 pKa = 5.33AVSFLLTYY428 pKa = 10.2GQTEE432 pKa = 4.29SVLPDD437 pKa = 3.1TGLSGFEE444 pKa = 3.68LVGAFRR450 pKa = 11.84DD451 pKa = 4.22GFLHH455 pKa = 7.16GGGGCDD461 pKa = 3.45VGLL464 pKa = 4.23
MM1 pKa = 7.35RR2 pKa = 11.84QVQRR6 pKa = 11.84RR7 pKa = 11.84ALAAAAAGSMLLAGCATTVTGTAAAAHH34 pKa = 7.06PLTDD38 pKa = 3.62VPAEE42 pKa = 4.01ALPITGAGDD51 pKa = 3.46GAIDD55 pKa = 3.6VLARR59 pKa = 11.84NAMADD64 pKa = 3.47LLTYY68 pKa = 9.05WEE70 pKa = 3.99RR71 pKa = 11.84TYY73 pKa = 11.11PEE75 pKa = 5.08LYY77 pKa = 10.65GEE79 pKa = 4.31PFVPLEE85 pKa = 4.1GGIFAVDD92 pKa = 4.58ADD94 pKa = 4.33DD95 pKa = 5.91LDD97 pKa = 4.22PAVYY101 pKa = 9.4PDD103 pKa = 3.83TGIGCDD109 pKa = 4.08RR110 pKa = 11.84LPVDD114 pKa = 3.61PSEE117 pKa = 4.46VEE119 pKa = 4.17GNAFYY124 pKa = 11.15LLPCDD129 pKa = 3.96LVAYY133 pKa = 10.05DD134 pKa = 3.62SALIEE139 pKa = 4.21EE140 pKa = 4.84LAAAHH145 pKa = 6.44GRR147 pKa = 11.84FLGPAVMAHH156 pKa = 5.74EE157 pKa = 5.05MGHH160 pKa = 7.21AIQGRR165 pKa = 11.84NGTWDD170 pKa = 3.43TVDD173 pKa = 3.88TIVSEE178 pKa = 4.33TQADD182 pKa = 4.12CFAGAWTRR190 pKa = 11.84WVADD194 pKa = 3.49GNAAHH199 pKa = 6.75SSLRR203 pKa = 11.84VPEE206 pKa = 4.26LDD208 pKa = 3.29EE209 pKa = 4.21VVVGFLEE216 pKa = 4.49LRR218 pKa = 11.84DD219 pKa = 3.84PVGTGTDD226 pKa = 3.02EE227 pKa = 5.08DD228 pKa = 4.62GAHH231 pKa = 6.62GSGFDD236 pKa = 3.23RR237 pKa = 11.84VSGFFAGWDD246 pKa = 3.63GGATACRR253 pKa = 11.84DD254 pKa = 3.5EE255 pKa = 4.53FDD257 pKa = 3.56EE258 pKa = 5.74DD259 pKa = 4.2RR260 pKa = 11.84LFTAAEE266 pKa = 4.17FTDD269 pKa = 4.53HH270 pKa = 7.29GDD272 pKa = 4.33LEE274 pKa = 4.73NEE276 pKa = 4.14GNASYY281 pKa = 11.35DD282 pKa = 3.57EE283 pKa = 4.21TLLIVDD289 pKa = 4.2ASLPLFYY296 pKa = 10.86EE297 pKa = 4.53SIFPSEE303 pKa = 3.97FGTRR307 pKa = 11.84FQVPALEE314 pKa = 4.9AFDD317 pKa = 4.48GTAPDD322 pKa = 4.0CGEE325 pKa = 4.39LGAHH329 pKa = 6.23GRR331 pKa = 11.84DD332 pKa = 3.5LGYY335 pKa = 10.6CGADD339 pKa = 3.0GTVYY343 pKa = 10.55FDD345 pKa = 3.51EE346 pKa = 4.98TDD348 pKa = 4.21LIEE351 pKa = 4.17PAYY354 pKa = 10.7AEE356 pKa = 4.58LGDD359 pKa = 3.78FAVATGIALPYY370 pKa = 10.52ALAARR375 pKa = 11.84DD376 pKa = 3.78QLGLSTDD383 pKa = 3.72DD384 pKa = 3.75GAATRR389 pKa = 11.84SAVCLTGWYY398 pKa = 7.87TADD401 pKa = 3.82FFAGDD406 pKa = 3.71FRR408 pKa = 11.84EE409 pKa = 4.5VTSLSPGDD417 pKa = 3.31VDD419 pKa = 3.7EE420 pKa = 5.33AVSFLLTYY428 pKa = 10.2GQTEE432 pKa = 4.29SVLPDD437 pKa = 3.1TGLSGFEE444 pKa = 3.68LVGAFRR450 pKa = 11.84DD451 pKa = 4.22GFLHH455 pKa = 7.16GGGGCDD461 pKa = 3.45VGLL464 pKa = 4.23
Molecular weight: 48.78 kDa
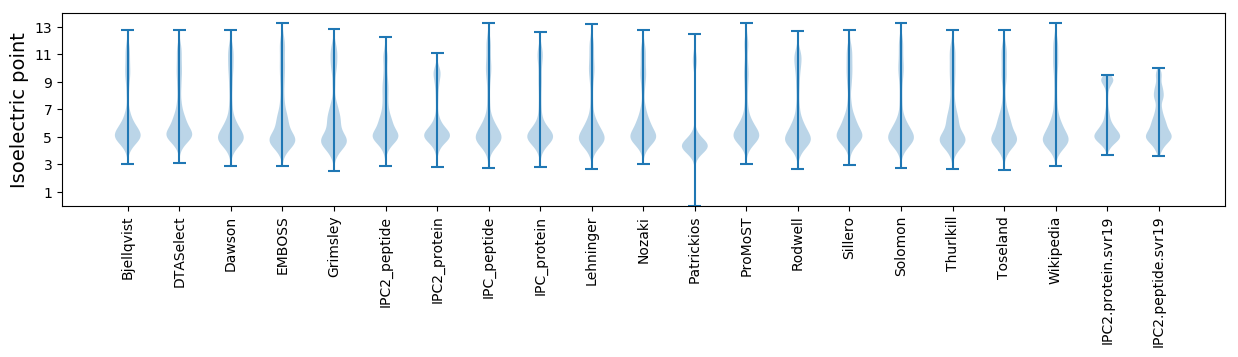
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3YRY7|A0A1I3YRY7_9ACTN Uncharacterized protein OS=Geodermatophilus ruber OX=504800 GN=SAMN04488085_101156 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
Molecular weight: 4.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1494515 |
29 |
2589 |
317.8 |
33.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.374 ± 0.056 | 0.709 ± 0.009 |
6.033 ± 0.027 | 5.747 ± 0.035 |
2.613 ± 0.023 | 9.623 ± 0.034 |
2.044 ± 0.016 | 2.981 ± 0.028 |
1.377 ± 0.023 | 10.704 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.644 ± 0.014 | 1.491 ± 0.018 |
6.378 ± 0.039 | 2.745 ± 0.021 |
8.289 ± 0.035 | 4.631 ± 0.021 |
5.83 ± 0.024 | 9.532 ± 0.039 |
1.461 ± 0.015 | 1.794 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
