
Anthurium mosaic-associated virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Chrysoviridae; Alphachrysovirus; Anthurium mosaic-associated chrysovirus
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
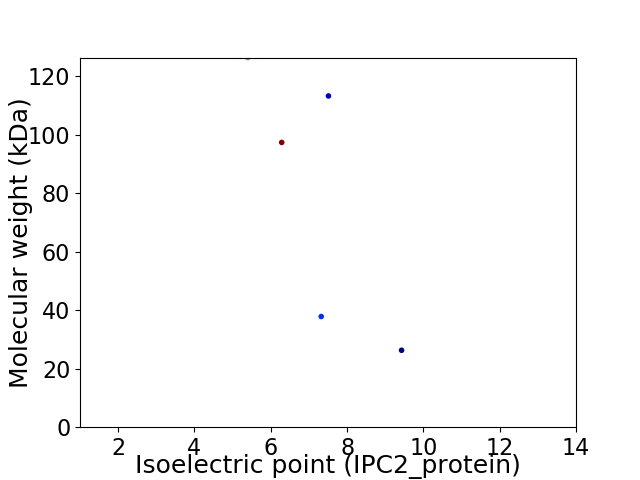
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
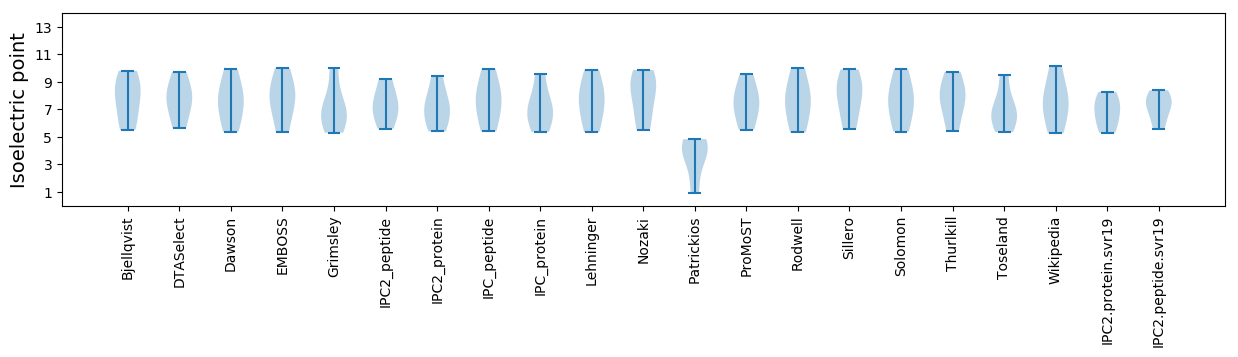
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D9U544|D9U544_9VIRU Coat protein OS=Anthurium mosaic-associated virus OX=664255 PE=4 SV=1
MM1 pKa = 7.99DD2 pKa = 5.08SEE4 pKa = 4.65IKK6 pKa = 10.33NDD8 pKa = 3.39SGRR11 pKa = 11.84FSADD15 pKa = 2.93KK16 pKa = 10.37IVSRR20 pKa = 11.84KK21 pKa = 9.06LRR23 pKa = 11.84AYY25 pKa = 10.44LGSTKK30 pKa = 9.68CTLEE34 pKa = 3.37YY35 pKa = 10.3RR36 pKa = 11.84EE37 pKa = 5.12RR38 pKa = 11.84IFSSLGRR45 pKa = 11.84LNDD48 pKa = 3.62KK49 pKa = 10.76VIEE52 pKa = 4.1GLYY55 pKa = 10.62AIVIPSGGGKK65 pKa = 6.03TTMARR70 pKa = 11.84EE71 pKa = 3.91FGFIDD76 pKa = 3.38VDD78 pKa = 3.86EE79 pKa = 5.24VIGNQAVMEE88 pKa = 4.24KK89 pKa = 10.44LVLEE93 pKa = 4.06RR94 pKa = 11.84RR95 pKa = 11.84EE96 pKa = 4.07LIRR99 pKa = 11.84WGKK102 pKa = 9.93DD103 pKa = 2.18RR104 pKa = 11.84WEE106 pKa = 4.02EE107 pKa = 4.19HH108 pKa = 5.62NKK110 pKa = 8.51VWYY113 pKa = 9.26NQVHH117 pKa = 5.63SVLAKK122 pKa = 10.53YY123 pKa = 10.18DD124 pKa = 3.61FQNNPGIIMVHH135 pKa = 4.74TEE137 pKa = 3.48EE138 pKa = 4.04MAYY141 pKa = 10.15EE142 pKa = 4.77LGAQPLIALTPKK154 pKa = 10.68DD155 pKa = 3.52SLYY158 pKa = 9.31KK159 pKa = 10.49QRR161 pKa = 11.84MKK163 pKa = 10.9GRR165 pKa = 11.84NEE167 pKa = 4.15LEE169 pKa = 3.77RR170 pKa = 11.84MLAVDD175 pKa = 3.07NLEE178 pKa = 4.18IVRR181 pKa = 11.84HH182 pKa = 5.06RR183 pKa = 11.84TQSVDD188 pKa = 2.99IIGYY192 pKa = 8.71GSWKK196 pKa = 9.81EE197 pKa = 3.91LKK199 pKa = 10.34QLIAGVAWEE208 pKa = 4.5EE209 pKa = 4.28VEE211 pKa = 4.04MAAPFRR217 pKa = 11.84DD218 pKa = 4.26YY219 pKa = 11.11NRR221 pKa = 11.84KK222 pKa = 8.11PAHH225 pKa = 6.21GWPIGYY231 pKa = 10.25GEE233 pKa = 5.8DD234 pKa = 3.25VALWIMQGKK243 pKa = 7.53TDD245 pKa = 4.11DD246 pKa = 4.01VEE248 pKa = 4.32DD249 pKa = 3.59MRR251 pKa = 11.84IVEE254 pKa = 4.12RR255 pKa = 11.84LHH257 pKa = 7.89RR258 pKa = 11.84EE259 pKa = 3.47GSVPDD264 pKa = 3.38ICLSYY269 pKa = 10.83YY270 pKa = 9.18SEE272 pKa = 4.75KK273 pKa = 10.39IYY275 pKa = 10.47GIKK278 pKa = 10.23KK279 pKa = 9.72IEE281 pKa = 4.34TKK283 pKa = 11.07GEE285 pKa = 4.12TLCRR289 pKa = 11.84EE290 pKa = 4.49WISICGEE297 pKa = 3.57ITNCGKK303 pKa = 9.62MDD305 pKa = 3.84TVTEE309 pKa = 4.18MEE311 pKa = 5.02VIANDD316 pKa = 3.61LNLVYY321 pKa = 9.93PYY323 pKa = 9.87EE324 pKa = 4.72SKK326 pKa = 10.23MAMNKK331 pKa = 8.36RR332 pKa = 11.84TIGLRR337 pKa = 11.84RR338 pKa = 11.84LLEE341 pKa = 4.08NMEE344 pKa = 4.18AMRR347 pKa = 11.84NDD349 pKa = 2.84IMRR352 pKa = 11.84TVLLNRR358 pKa = 11.84RR359 pKa = 11.84GSNQNLVVGILIWVGGVLSNMRR381 pKa = 11.84VEE383 pKa = 4.38IQDD386 pKa = 3.32SVLRR390 pKa = 11.84SEE392 pKa = 4.5ILLVPDD398 pKa = 3.89EE399 pKa = 5.08DD400 pKa = 4.17WIKK403 pKa = 10.25MGKK406 pKa = 9.14KK407 pKa = 8.2VHH409 pKa = 6.72DD410 pKa = 3.63MVRR413 pKa = 11.84RR414 pKa = 11.84TGTFFGTTINVQEE427 pKa = 4.35AQKK430 pKa = 11.21LMYY433 pKa = 10.91LHH435 pKa = 6.34MLYY438 pKa = 10.79GRR440 pKa = 11.84FVYY443 pKa = 10.28QVDD446 pKa = 3.77EE447 pKa = 4.1EE448 pKa = 4.58AEE450 pKa = 3.54IDD452 pKa = 3.45KK453 pKa = 10.77RR454 pKa = 11.84KK455 pKa = 10.31RR456 pKa = 11.84EE457 pKa = 4.05LNDD460 pKa = 2.87TKK462 pKa = 10.34MAYY465 pKa = 10.38LDD467 pKa = 3.89GEE469 pKa = 4.26WRR471 pKa = 11.84VDD473 pKa = 3.49EE474 pKa = 4.24FDD476 pKa = 3.65KK477 pKa = 11.75LEE479 pKa = 4.3LEE481 pKa = 5.06GIRR484 pKa = 11.84DD485 pKa = 3.98AYY487 pKa = 10.77RR488 pKa = 11.84RR489 pKa = 11.84LCDD492 pKa = 3.58SITPQRR498 pKa = 11.84VMDD501 pKa = 5.42FSEE504 pKa = 4.31FWKK507 pKa = 10.44RR508 pKa = 11.84RR509 pKa = 11.84RR510 pKa = 11.84SWAAKK515 pKa = 9.0GSTVVYY521 pKa = 9.77EE522 pKa = 4.3GEE524 pKa = 4.11KK525 pKa = 10.44KK526 pKa = 10.83YY527 pKa = 10.31MLEE530 pKa = 3.88IVDD533 pKa = 3.65EE534 pKa = 4.78VIRR537 pKa = 11.84QLEE540 pKa = 3.94LRR542 pKa = 11.84HH543 pKa = 5.71NKK545 pKa = 9.28KK546 pKa = 10.67SLMEE550 pKa = 5.48DD551 pKa = 3.13EE552 pKa = 5.54AEE554 pKa = 4.12CCKK557 pKa = 10.29IISGIIRR564 pKa = 11.84DD565 pKa = 4.15LGRR568 pKa = 11.84NDD570 pKa = 3.0TKK572 pKa = 10.41MVPKK576 pKa = 10.38FEE578 pKa = 4.46SAAKK582 pKa = 10.01RR583 pKa = 11.84ALLPGNLYY591 pKa = 10.26HH592 pKa = 6.49YY593 pKa = 10.89VVFSYY598 pKa = 11.37VLLIFEE604 pKa = 4.66NCASIGDD611 pKa = 3.76VRR613 pKa = 11.84LGNDD617 pKa = 3.2RR618 pKa = 11.84DD619 pKa = 3.73NSFGAFDD626 pKa = 4.82SKK628 pKa = 11.58LEE630 pKa = 4.19TNLTRR635 pKa = 11.84FVYY638 pKa = 10.64DD639 pKa = 3.72FADD642 pKa = 4.83FNAQHH647 pKa = 6.34SRR649 pKa = 11.84RR650 pKa = 11.84SMARR654 pKa = 11.84VMSVLSEE661 pKa = 4.06TVDD664 pKa = 3.56PNEE667 pKa = 4.05TLGFCLKK674 pKa = 9.93WMSLSFNKK682 pKa = 9.79MEE684 pKa = 4.45VIKK687 pKa = 10.84EE688 pKa = 4.0GGGVEE693 pKa = 4.11KK694 pKa = 10.56LKK696 pKa = 11.06SGLYY700 pKa = 9.77SGWRR704 pKa = 11.84GTTWINSVLNHH715 pKa = 6.59AYY717 pKa = 9.09MYY719 pKa = 9.67VARR722 pKa = 11.84VCFNRR727 pKa = 11.84LYY729 pKa = 10.52GYY731 pKa = 10.33EE732 pKa = 4.07PFVEE736 pKa = 4.22YY737 pKa = 10.46EE738 pKa = 4.15GAGDD742 pKa = 4.31DD743 pKa = 3.49VDD745 pKa = 5.49GVVKK749 pKa = 10.64DD750 pKa = 3.36ISTAGKK756 pKa = 9.65LYY758 pKa = 10.54RR759 pKa = 11.84ITVEE763 pKa = 4.49MGLEE767 pKa = 4.2SNVQKK772 pKa = 10.9QLFGKK777 pKa = 9.72RR778 pKa = 11.84AEE780 pKa = 4.17FLRR783 pKa = 11.84VSYY786 pKa = 10.57EE787 pKa = 3.58SGYY790 pKa = 10.94AGSSICRR797 pKa = 11.84VLGNFISGNWEE808 pKa = 3.95GEE810 pKa = 4.2GGSVSDD816 pKa = 4.56RR817 pKa = 11.84LTSAIDD823 pKa = 3.72NILTLGRR830 pKa = 11.84RR831 pKa = 11.84GLDD834 pKa = 2.95EE835 pKa = 5.52SMVKK839 pKa = 10.03VLYY842 pKa = 10.26RR843 pKa = 11.84CVLMHH848 pKa = 6.66VGRR851 pKa = 11.84IFDD854 pKa = 3.69GDD856 pKa = 3.31EE857 pKa = 4.15WIGLSPCILHH867 pKa = 7.09GRR869 pKa = 11.84MEE871 pKa = 5.14DD872 pKa = 3.34NGFGIPDD879 pKa = 3.35EE880 pKa = 5.23DD881 pKa = 3.96GCVWEE886 pKa = 4.97LEE888 pKa = 4.49KK889 pKa = 10.84KK890 pKa = 10.72APAPDD895 pKa = 3.03GWLGYY900 pKa = 9.99IKK902 pKa = 10.66LPAKK906 pKa = 8.88TASRR910 pKa = 11.84DD911 pKa = 3.33WVDD914 pKa = 4.26VIDD917 pKa = 4.19EE918 pKa = 4.55EE919 pKa = 5.07IRR921 pKa = 11.84DD922 pKa = 3.76RR923 pKa = 11.84GLKK926 pKa = 10.35VGDD929 pKa = 3.43VDD931 pKa = 5.13RR932 pKa = 11.84LSEE935 pKa = 4.0LLARR939 pKa = 11.84DD940 pKa = 3.66SYY942 pKa = 11.25DD943 pKa = 3.32VKK945 pKa = 11.11SVADD949 pKa = 3.89RR950 pKa = 11.84FGTEE954 pKa = 3.69ISPEE958 pKa = 3.36IWKK961 pKa = 10.29KK962 pKa = 9.44YY963 pKa = 4.74WTYY966 pKa = 10.16KK967 pKa = 10.09CKK969 pKa = 10.45VVKK972 pKa = 10.05KK973 pKa = 9.65YY974 pKa = 10.64KK975 pKa = 10.21IEE977 pKa = 3.93TDD979 pKa = 3.75VIDD982 pKa = 4.39EE983 pKa = 4.3GMLLDD988 pKa = 3.84FLEE991 pKa = 4.31WLGGEE996 pKa = 4.14EE997 pKa = 4.94TPTLDD1002 pKa = 3.92QLSKK1006 pKa = 11.55LEE1008 pKa = 3.81ILGPYY1013 pKa = 9.64IEE1015 pKa = 5.26TIVSGDD1021 pKa = 3.68DD1022 pKa = 3.4KK1023 pKa = 11.2PVTEE1027 pKa = 4.31EE1028 pKa = 3.3EE1029 pKa = 4.97WYY1031 pKa = 11.06GEE1033 pKa = 4.51LYY1035 pKa = 10.4DD1036 pKa = 4.5APRR1039 pKa = 11.84RR1040 pKa = 11.84LEE1042 pKa = 3.87NMKK1045 pKa = 9.79LQRR1048 pKa = 11.84WKK1050 pKa = 11.08VVLCPPLVQSKK1061 pKa = 9.48IARR1064 pKa = 11.84WCKK1067 pKa = 10.35EE1068 pKa = 3.34MVANDD1073 pKa = 3.69MVTQFTGMRR1082 pKa = 11.84IYY1084 pKa = 10.66QIVSNTYY1091 pKa = 8.33GSIFGFEE1098 pKa = 3.85II1099 pKa = 4.17
MM1 pKa = 7.99DD2 pKa = 5.08SEE4 pKa = 4.65IKK6 pKa = 10.33NDD8 pKa = 3.39SGRR11 pKa = 11.84FSADD15 pKa = 2.93KK16 pKa = 10.37IVSRR20 pKa = 11.84KK21 pKa = 9.06LRR23 pKa = 11.84AYY25 pKa = 10.44LGSTKK30 pKa = 9.68CTLEE34 pKa = 3.37YY35 pKa = 10.3RR36 pKa = 11.84EE37 pKa = 5.12RR38 pKa = 11.84IFSSLGRR45 pKa = 11.84LNDD48 pKa = 3.62KK49 pKa = 10.76VIEE52 pKa = 4.1GLYY55 pKa = 10.62AIVIPSGGGKK65 pKa = 6.03TTMARR70 pKa = 11.84EE71 pKa = 3.91FGFIDD76 pKa = 3.38VDD78 pKa = 3.86EE79 pKa = 5.24VIGNQAVMEE88 pKa = 4.24KK89 pKa = 10.44LVLEE93 pKa = 4.06RR94 pKa = 11.84RR95 pKa = 11.84EE96 pKa = 4.07LIRR99 pKa = 11.84WGKK102 pKa = 9.93DD103 pKa = 2.18RR104 pKa = 11.84WEE106 pKa = 4.02EE107 pKa = 4.19HH108 pKa = 5.62NKK110 pKa = 8.51VWYY113 pKa = 9.26NQVHH117 pKa = 5.63SVLAKK122 pKa = 10.53YY123 pKa = 10.18DD124 pKa = 3.61FQNNPGIIMVHH135 pKa = 4.74TEE137 pKa = 3.48EE138 pKa = 4.04MAYY141 pKa = 10.15EE142 pKa = 4.77LGAQPLIALTPKK154 pKa = 10.68DD155 pKa = 3.52SLYY158 pKa = 9.31KK159 pKa = 10.49QRR161 pKa = 11.84MKK163 pKa = 10.9GRR165 pKa = 11.84NEE167 pKa = 4.15LEE169 pKa = 3.77RR170 pKa = 11.84MLAVDD175 pKa = 3.07NLEE178 pKa = 4.18IVRR181 pKa = 11.84HH182 pKa = 5.06RR183 pKa = 11.84TQSVDD188 pKa = 2.99IIGYY192 pKa = 8.71GSWKK196 pKa = 9.81EE197 pKa = 3.91LKK199 pKa = 10.34QLIAGVAWEE208 pKa = 4.5EE209 pKa = 4.28VEE211 pKa = 4.04MAAPFRR217 pKa = 11.84DD218 pKa = 4.26YY219 pKa = 11.11NRR221 pKa = 11.84KK222 pKa = 8.11PAHH225 pKa = 6.21GWPIGYY231 pKa = 10.25GEE233 pKa = 5.8DD234 pKa = 3.25VALWIMQGKK243 pKa = 7.53TDD245 pKa = 4.11DD246 pKa = 4.01VEE248 pKa = 4.32DD249 pKa = 3.59MRR251 pKa = 11.84IVEE254 pKa = 4.12RR255 pKa = 11.84LHH257 pKa = 7.89RR258 pKa = 11.84EE259 pKa = 3.47GSVPDD264 pKa = 3.38ICLSYY269 pKa = 10.83YY270 pKa = 9.18SEE272 pKa = 4.75KK273 pKa = 10.39IYY275 pKa = 10.47GIKK278 pKa = 10.23KK279 pKa = 9.72IEE281 pKa = 4.34TKK283 pKa = 11.07GEE285 pKa = 4.12TLCRR289 pKa = 11.84EE290 pKa = 4.49WISICGEE297 pKa = 3.57ITNCGKK303 pKa = 9.62MDD305 pKa = 3.84TVTEE309 pKa = 4.18MEE311 pKa = 5.02VIANDD316 pKa = 3.61LNLVYY321 pKa = 9.93PYY323 pKa = 9.87EE324 pKa = 4.72SKK326 pKa = 10.23MAMNKK331 pKa = 8.36RR332 pKa = 11.84TIGLRR337 pKa = 11.84RR338 pKa = 11.84LLEE341 pKa = 4.08NMEE344 pKa = 4.18AMRR347 pKa = 11.84NDD349 pKa = 2.84IMRR352 pKa = 11.84TVLLNRR358 pKa = 11.84RR359 pKa = 11.84GSNQNLVVGILIWVGGVLSNMRR381 pKa = 11.84VEE383 pKa = 4.38IQDD386 pKa = 3.32SVLRR390 pKa = 11.84SEE392 pKa = 4.5ILLVPDD398 pKa = 3.89EE399 pKa = 5.08DD400 pKa = 4.17WIKK403 pKa = 10.25MGKK406 pKa = 9.14KK407 pKa = 8.2VHH409 pKa = 6.72DD410 pKa = 3.63MVRR413 pKa = 11.84RR414 pKa = 11.84TGTFFGTTINVQEE427 pKa = 4.35AQKK430 pKa = 11.21LMYY433 pKa = 10.91LHH435 pKa = 6.34MLYY438 pKa = 10.79GRR440 pKa = 11.84FVYY443 pKa = 10.28QVDD446 pKa = 3.77EE447 pKa = 4.1EE448 pKa = 4.58AEE450 pKa = 3.54IDD452 pKa = 3.45KK453 pKa = 10.77RR454 pKa = 11.84KK455 pKa = 10.31RR456 pKa = 11.84EE457 pKa = 4.05LNDD460 pKa = 2.87TKK462 pKa = 10.34MAYY465 pKa = 10.38LDD467 pKa = 3.89GEE469 pKa = 4.26WRR471 pKa = 11.84VDD473 pKa = 3.49EE474 pKa = 4.24FDD476 pKa = 3.65KK477 pKa = 11.75LEE479 pKa = 4.3LEE481 pKa = 5.06GIRR484 pKa = 11.84DD485 pKa = 3.98AYY487 pKa = 10.77RR488 pKa = 11.84RR489 pKa = 11.84LCDD492 pKa = 3.58SITPQRR498 pKa = 11.84VMDD501 pKa = 5.42FSEE504 pKa = 4.31FWKK507 pKa = 10.44RR508 pKa = 11.84RR509 pKa = 11.84RR510 pKa = 11.84SWAAKK515 pKa = 9.0GSTVVYY521 pKa = 9.77EE522 pKa = 4.3GEE524 pKa = 4.11KK525 pKa = 10.44KK526 pKa = 10.83YY527 pKa = 10.31MLEE530 pKa = 3.88IVDD533 pKa = 3.65EE534 pKa = 4.78VIRR537 pKa = 11.84QLEE540 pKa = 3.94LRR542 pKa = 11.84HH543 pKa = 5.71NKK545 pKa = 9.28KK546 pKa = 10.67SLMEE550 pKa = 5.48DD551 pKa = 3.13EE552 pKa = 5.54AEE554 pKa = 4.12CCKK557 pKa = 10.29IISGIIRR564 pKa = 11.84DD565 pKa = 4.15LGRR568 pKa = 11.84NDD570 pKa = 3.0TKK572 pKa = 10.41MVPKK576 pKa = 10.38FEE578 pKa = 4.46SAAKK582 pKa = 10.01RR583 pKa = 11.84ALLPGNLYY591 pKa = 10.26HH592 pKa = 6.49YY593 pKa = 10.89VVFSYY598 pKa = 11.37VLLIFEE604 pKa = 4.66NCASIGDD611 pKa = 3.76VRR613 pKa = 11.84LGNDD617 pKa = 3.2RR618 pKa = 11.84DD619 pKa = 3.73NSFGAFDD626 pKa = 4.82SKK628 pKa = 11.58LEE630 pKa = 4.19TNLTRR635 pKa = 11.84FVYY638 pKa = 10.64DD639 pKa = 3.72FADD642 pKa = 4.83FNAQHH647 pKa = 6.34SRR649 pKa = 11.84RR650 pKa = 11.84SMARR654 pKa = 11.84VMSVLSEE661 pKa = 4.06TVDD664 pKa = 3.56PNEE667 pKa = 4.05TLGFCLKK674 pKa = 9.93WMSLSFNKK682 pKa = 9.79MEE684 pKa = 4.45VIKK687 pKa = 10.84EE688 pKa = 4.0GGGVEE693 pKa = 4.11KK694 pKa = 10.56LKK696 pKa = 11.06SGLYY700 pKa = 9.77SGWRR704 pKa = 11.84GTTWINSVLNHH715 pKa = 6.59AYY717 pKa = 9.09MYY719 pKa = 9.67VARR722 pKa = 11.84VCFNRR727 pKa = 11.84LYY729 pKa = 10.52GYY731 pKa = 10.33EE732 pKa = 4.07PFVEE736 pKa = 4.22YY737 pKa = 10.46EE738 pKa = 4.15GAGDD742 pKa = 4.31DD743 pKa = 3.49VDD745 pKa = 5.49GVVKK749 pKa = 10.64DD750 pKa = 3.36ISTAGKK756 pKa = 9.65LYY758 pKa = 10.54RR759 pKa = 11.84ITVEE763 pKa = 4.49MGLEE767 pKa = 4.2SNVQKK772 pKa = 10.9QLFGKK777 pKa = 9.72RR778 pKa = 11.84AEE780 pKa = 4.17FLRR783 pKa = 11.84VSYY786 pKa = 10.57EE787 pKa = 3.58SGYY790 pKa = 10.94AGSSICRR797 pKa = 11.84VLGNFISGNWEE808 pKa = 3.95GEE810 pKa = 4.2GGSVSDD816 pKa = 4.56RR817 pKa = 11.84LTSAIDD823 pKa = 3.72NILTLGRR830 pKa = 11.84RR831 pKa = 11.84GLDD834 pKa = 2.95EE835 pKa = 5.52SMVKK839 pKa = 10.03VLYY842 pKa = 10.26RR843 pKa = 11.84CVLMHH848 pKa = 6.66VGRR851 pKa = 11.84IFDD854 pKa = 3.69GDD856 pKa = 3.31EE857 pKa = 4.15WIGLSPCILHH867 pKa = 7.09GRR869 pKa = 11.84MEE871 pKa = 5.14DD872 pKa = 3.34NGFGIPDD879 pKa = 3.35EE880 pKa = 5.23DD881 pKa = 3.96GCVWEE886 pKa = 4.97LEE888 pKa = 4.49KK889 pKa = 10.84KK890 pKa = 10.72APAPDD895 pKa = 3.03GWLGYY900 pKa = 9.99IKK902 pKa = 10.66LPAKK906 pKa = 8.88TASRR910 pKa = 11.84DD911 pKa = 3.33WVDD914 pKa = 4.26VIDD917 pKa = 4.19EE918 pKa = 4.55EE919 pKa = 5.07IRR921 pKa = 11.84DD922 pKa = 3.76RR923 pKa = 11.84GLKK926 pKa = 10.35VGDD929 pKa = 3.43VDD931 pKa = 5.13RR932 pKa = 11.84LSEE935 pKa = 4.0LLARR939 pKa = 11.84DD940 pKa = 3.66SYY942 pKa = 11.25DD943 pKa = 3.32VKK945 pKa = 11.11SVADD949 pKa = 3.89RR950 pKa = 11.84FGTEE954 pKa = 3.69ISPEE958 pKa = 3.36IWKK961 pKa = 10.29KK962 pKa = 9.44YY963 pKa = 4.74WTYY966 pKa = 10.16KK967 pKa = 10.09CKK969 pKa = 10.45VVKK972 pKa = 10.05KK973 pKa = 9.65YY974 pKa = 10.64KK975 pKa = 10.21IEE977 pKa = 3.93TDD979 pKa = 3.75VIDD982 pKa = 4.39EE983 pKa = 4.3GMLLDD988 pKa = 3.84FLEE991 pKa = 4.31WLGGEE996 pKa = 4.14EE997 pKa = 4.94TPTLDD1002 pKa = 3.92QLSKK1006 pKa = 11.55LEE1008 pKa = 3.81ILGPYY1013 pKa = 9.64IEE1015 pKa = 5.26TIVSGDD1021 pKa = 3.68DD1022 pKa = 3.4KK1023 pKa = 11.2PVTEE1027 pKa = 4.31EE1028 pKa = 3.3EE1029 pKa = 4.97WYY1031 pKa = 11.06GEE1033 pKa = 4.51LYY1035 pKa = 10.4DD1036 pKa = 4.5APRR1039 pKa = 11.84RR1040 pKa = 11.84LEE1042 pKa = 3.87NMKK1045 pKa = 9.79LQRR1048 pKa = 11.84WKK1050 pKa = 11.08VVLCPPLVQSKK1061 pKa = 9.48IARR1064 pKa = 11.84WCKK1067 pKa = 10.35EE1068 pKa = 3.34MVANDD1073 pKa = 3.69MVTQFTGMRR1082 pKa = 11.84IYY1084 pKa = 10.66QIVSNTYY1091 pKa = 8.33GSIFGFEE1098 pKa = 3.85II1099 pKa = 4.17
Molecular weight: 126.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D9U547|D9U547_9VIRU p26 OS=Anthurium mosaic-associated virus OX=664255 PE=4 SV=1
MM1 pKa = 6.31TTPYY5 pKa = 10.36PRR7 pKa = 11.84FAINRR12 pKa = 11.84AAGSISNQSFFSCSLLKK29 pKa = 10.95YY30 pKa = 8.67STNSLPVRR38 pKa = 11.84LCPCSLRR45 pKa = 11.84PSHH48 pKa = 6.36NAWATHH54 pKa = 5.73NPSPTTCFTFSFVPSAVTSLMNVSIPLNPKK84 pKa = 9.87LSAIFLMKK92 pKa = 10.66LIFGGFLAFRR102 pKa = 11.84PPLFNLSVASMSSSSLLSFPFSGFTPASAFVFSFLLSPLFCLSSFSILSSRR153 pKa = 11.84SLCALSFCSLLSFSIPRR170 pKa = 11.84YY171 pKa = 8.32VLKK174 pKa = 10.68KK175 pKa = 10.29SNLSFSIVSLPTLGSYY191 pKa = 10.93SSFQFVSRR199 pKa = 11.84YY200 pKa = 6.67TSSYY204 pKa = 8.75TPYY207 pKa = 9.61PPSILAIVLWLTISSPIFCIALYY230 pKa = 8.18PQNIIISSLSPP241 pKa = 3.44
MM1 pKa = 6.31TTPYY5 pKa = 10.36PRR7 pKa = 11.84FAINRR12 pKa = 11.84AAGSISNQSFFSCSLLKK29 pKa = 10.95YY30 pKa = 8.67STNSLPVRR38 pKa = 11.84LCPCSLRR45 pKa = 11.84PSHH48 pKa = 6.36NAWATHH54 pKa = 5.73NPSPTTCFTFSFVPSAVTSLMNVSIPLNPKK84 pKa = 9.87LSAIFLMKK92 pKa = 10.66LIFGGFLAFRR102 pKa = 11.84PPLFNLSVASMSSSSLLSFPFSGFTPASAFVFSFLLSPLFCLSSFSILSSRR153 pKa = 11.84SLCALSFCSLLSFSIPRR170 pKa = 11.84YY171 pKa = 8.32VLKK174 pKa = 10.68KK175 pKa = 10.29SNLSFSIVSLPTLGSYY191 pKa = 10.93SSFQFVSRR199 pKa = 11.84YY200 pKa = 6.67TSSYY204 pKa = 8.75TPYY207 pKa = 9.61PPSILAIVLWLTISSPIFCIALYY230 pKa = 8.18PQNIIISSLSPP241 pKa = 3.44
Molecular weight: 26.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3544 |
241 |
1099 |
708.8 |
80.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.91 ± 0.421 | 2.088 ± 0.267 |
4.853 ± 0.898 | 8.465 ± 1.922 |
4.007 ± 1.441 | 7.562 ± 1.287 |
1.467 ± 0.224 | 6.405 ± 0.279 |
7.336 ± 1.319 | 9.255 ± 1.277 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.386 ± 0.375 | 4.374 ± 0.491 |
3.612 ± 1.218 | 1.862 ± 0.226 |
6.01 ± 0.922 | 8.775 ± 2.653 |
4.402 ± 0.221 | 6.123 ± 0.714 |
2.088 ± 0.252 | 3.019 ± 0.547 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
