
Salegentibacter flavus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Salegentibacter
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
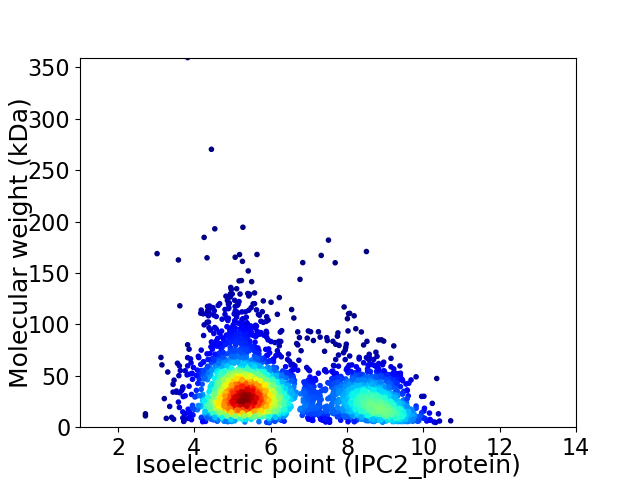
Virtual 2D-PAGE plot for 3319 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4Z736|A0A1I4Z736_9FLAO Predicted N-formylglutamate amidohydrolase OS=Salegentibacter flavus OX=287099 GN=SAMN05660413_01154 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 10.38NILKK6 pKa = 10.24ISLSVFMLVFFMACTDD22 pKa = 3.61DD23 pKa = 5.77DD24 pKa = 4.76YY25 pKa = 11.76DD26 pKa = 3.91IGEE29 pKa = 4.16ITAPTNLQVEE39 pKa = 4.54TAILGQSDD47 pKa = 4.55DD48 pKa = 4.26MPDD51 pKa = 3.44GDD53 pKa = 4.43GSGEE57 pKa = 4.15VTFTATAQGAMTYY70 pKa = 10.9KK71 pKa = 10.84FIFEE75 pKa = 4.64NGTEE79 pKa = 4.12VTTASGVYY87 pKa = 7.04THH89 pKa = 7.43PFSEE93 pKa = 4.82TGTQTYY99 pKa = 9.51NVNIIAYY106 pKa = 8.72GPGGTASSTSVEE118 pKa = 4.22VAVLVTYY125 pKa = 7.85EE126 pKa = 4.22PPADD130 pKa = 4.04LLEE133 pKa = 4.22KK134 pKa = 10.8LIGDD138 pKa = 4.16GAKK141 pKa = 9.31EE142 pKa = 3.75WRR144 pKa = 11.84IKK146 pKa = 11.01AEE148 pKa = 3.97AAGHH152 pKa = 6.22FGLGPVGGSIPTEE165 pKa = 4.0WYY167 pKa = 10.16GAGPNEE173 pKa = 4.18KK174 pKa = 9.78AHH176 pKa = 6.14TGMYY180 pKa = 9.3DD181 pKa = 3.04DD182 pKa = 5.6RR183 pKa = 11.84YY184 pKa = 10.48IFEE187 pKa = 4.31EE188 pKa = 4.95DD189 pKa = 3.29GTFVHH194 pKa = 6.49ITDD197 pKa = 3.9ITNDD201 pKa = 3.81DD202 pKa = 3.27ASGTVFGRR210 pKa = 11.84APLVEE215 pKa = 4.07QLGVSCDD222 pKa = 3.83CEE224 pKa = 4.46TEE226 pKa = 4.11GDD228 pKa = 3.88DD229 pKa = 5.18VLNIPYY235 pKa = 10.26DD236 pKa = 4.03DD237 pKa = 3.93YY238 pKa = 11.6TEE240 pKa = 4.02NWSISAPGGNEE251 pKa = 4.06TINLTGIGFIGYY263 pKa = 9.72YY264 pKa = 9.54IGGDD268 pKa = 3.15HH269 pKa = 7.24RR270 pKa = 11.84YY271 pKa = 9.84EE272 pKa = 3.93IFDD275 pKa = 3.29RR276 pKa = 11.84SVPNEE281 pKa = 4.35LIIKK285 pKa = 8.38STDD288 pKa = 3.03GNGDD292 pKa = 3.55FDD294 pKa = 3.22WWFILTSDD302 pKa = 3.63SGGGAEE308 pKa = 4.18EE309 pKa = 4.5EE310 pKa = 4.43FVSEE314 pKa = 4.0YY315 pKa = 11.28NEE317 pKa = 3.69LHH319 pKa = 6.88RR320 pKa = 11.84EE321 pKa = 4.06FNFDD325 pKa = 3.25TDD327 pKa = 4.12GEE329 pKa = 4.63LDD331 pKa = 3.48TTVWNFEE338 pKa = 4.24TGNGEE343 pKa = 4.44DD344 pKa = 3.07GWGNQEE350 pKa = 3.79SQYY353 pKa = 9.2YY354 pKa = 8.98TEE356 pKa = 5.07DD357 pKa = 3.29NAVISGGNLVITAKK371 pKa = 10.94AEE373 pKa = 4.6DD374 pKa = 3.96IEE376 pKa = 4.68GFDD379 pKa = 3.92YY380 pKa = 10.93SSSRR384 pKa = 11.84ITTKK388 pKa = 11.12DD389 pKa = 3.0NFEE392 pKa = 3.94FTYY395 pKa = 10.88GRR397 pKa = 11.84IEE399 pKa = 4.03ARR401 pKa = 11.84AKK403 pKa = 10.46LPEE406 pKa = 4.56GAGTWPAIWMLGSDD420 pKa = 4.11FDD422 pKa = 4.48EE423 pKa = 4.92VGWPQTGEE431 pKa = 3.59IDD433 pKa = 3.93IMEE436 pKa = 4.59HH437 pKa = 6.73AGNEE441 pKa = 3.57QDD443 pKa = 5.22LIHH446 pKa = 6.44GTLHH450 pKa = 5.72YY451 pKa = 9.82EE452 pKa = 4.17GRR454 pKa = 11.84SGGDD458 pKa = 2.85ADD460 pKa = 4.22GNTIEE465 pKa = 4.37VDD467 pKa = 3.65GVSDD471 pKa = 4.09DD472 pKa = 3.57FHH474 pKa = 8.72IYY476 pKa = 7.95TVEE479 pKa = 3.65WSDD482 pKa = 3.12EE483 pKa = 4.32HH484 pKa = 9.22IIFLVDD490 pKa = 3.15GEE492 pKa = 4.59VFHH495 pKa = 6.89TYY497 pKa = 10.32EE498 pKa = 4.69NNPDD502 pKa = 3.39SPFNKK507 pKa = 10.16DD508 pKa = 3.31FFLILNVAMGGTFGGEE524 pKa = 3.72INPDD528 pKa = 3.47FVEE531 pKa = 4.47SSMEE535 pKa = 3.39VDD537 pKa = 4.17YY538 pKa = 11.42IRR540 pKa = 11.84VFQQ543 pKa = 4.13
MM1 pKa = 7.51KK2 pKa = 10.38NILKK6 pKa = 10.24ISLSVFMLVFFMACTDD22 pKa = 3.61DD23 pKa = 5.77DD24 pKa = 4.76YY25 pKa = 11.76DD26 pKa = 3.91IGEE29 pKa = 4.16ITAPTNLQVEE39 pKa = 4.54TAILGQSDD47 pKa = 4.55DD48 pKa = 4.26MPDD51 pKa = 3.44GDD53 pKa = 4.43GSGEE57 pKa = 4.15VTFTATAQGAMTYY70 pKa = 10.9KK71 pKa = 10.84FIFEE75 pKa = 4.64NGTEE79 pKa = 4.12VTTASGVYY87 pKa = 7.04THH89 pKa = 7.43PFSEE93 pKa = 4.82TGTQTYY99 pKa = 9.51NVNIIAYY106 pKa = 8.72GPGGTASSTSVEE118 pKa = 4.22VAVLVTYY125 pKa = 7.85EE126 pKa = 4.22PPADD130 pKa = 4.04LLEE133 pKa = 4.22KK134 pKa = 10.8LIGDD138 pKa = 4.16GAKK141 pKa = 9.31EE142 pKa = 3.75WRR144 pKa = 11.84IKK146 pKa = 11.01AEE148 pKa = 3.97AAGHH152 pKa = 6.22FGLGPVGGSIPTEE165 pKa = 4.0WYY167 pKa = 10.16GAGPNEE173 pKa = 4.18KK174 pKa = 9.78AHH176 pKa = 6.14TGMYY180 pKa = 9.3DD181 pKa = 3.04DD182 pKa = 5.6RR183 pKa = 11.84YY184 pKa = 10.48IFEE187 pKa = 4.31EE188 pKa = 4.95DD189 pKa = 3.29GTFVHH194 pKa = 6.49ITDD197 pKa = 3.9ITNDD201 pKa = 3.81DD202 pKa = 3.27ASGTVFGRR210 pKa = 11.84APLVEE215 pKa = 4.07QLGVSCDD222 pKa = 3.83CEE224 pKa = 4.46TEE226 pKa = 4.11GDD228 pKa = 3.88DD229 pKa = 5.18VLNIPYY235 pKa = 10.26DD236 pKa = 4.03DD237 pKa = 3.93YY238 pKa = 11.6TEE240 pKa = 4.02NWSISAPGGNEE251 pKa = 4.06TINLTGIGFIGYY263 pKa = 9.72YY264 pKa = 9.54IGGDD268 pKa = 3.15HH269 pKa = 7.24RR270 pKa = 11.84YY271 pKa = 9.84EE272 pKa = 3.93IFDD275 pKa = 3.29RR276 pKa = 11.84SVPNEE281 pKa = 4.35LIIKK285 pKa = 8.38STDD288 pKa = 3.03GNGDD292 pKa = 3.55FDD294 pKa = 3.22WWFILTSDD302 pKa = 3.63SGGGAEE308 pKa = 4.18EE309 pKa = 4.5EE310 pKa = 4.43FVSEE314 pKa = 4.0YY315 pKa = 11.28NEE317 pKa = 3.69LHH319 pKa = 6.88RR320 pKa = 11.84EE321 pKa = 4.06FNFDD325 pKa = 3.25TDD327 pKa = 4.12GEE329 pKa = 4.63LDD331 pKa = 3.48TTVWNFEE338 pKa = 4.24TGNGEE343 pKa = 4.44DD344 pKa = 3.07GWGNQEE350 pKa = 3.79SQYY353 pKa = 9.2YY354 pKa = 8.98TEE356 pKa = 5.07DD357 pKa = 3.29NAVISGGNLVITAKK371 pKa = 10.94AEE373 pKa = 4.6DD374 pKa = 3.96IEE376 pKa = 4.68GFDD379 pKa = 3.92YY380 pKa = 10.93SSSRR384 pKa = 11.84ITTKK388 pKa = 11.12DD389 pKa = 3.0NFEE392 pKa = 3.94FTYY395 pKa = 10.88GRR397 pKa = 11.84IEE399 pKa = 4.03ARR401 pKa = 11.84AKK403 pKa = 10.46LPEE406 pKa = 4.56GAGTWPAIWMLGSDD420 pKa = 4.11FDD422 pKa = 4.48EE423 pKa = 4.92VGWPQTGEE431 pKa = 3.59IDD433 pKa = 3.93IMEE436 pKa = 4.59HH437 pKa = 6.73AGNEE441 pKa = 3.57QDD443 pKa = 5.22LIHH446 pKa = 6.44GTLHH450 pKa = 5.72YY451 pKa = 9.82EE452 pKa = 4.17GRR454 pKa = 11.84SGGDD458 pKa = 2.85ADD460 pKa = 4.22GNTIEE465 pKa = 4.37VDD467 pKa = 3.65GVSDD471 pKa = 4.09DD472 pKa = 3.57FHH474 pKa = 8.72IYY476 pKa = 7.95TVEE479 pKa = 3.65WSDD482 pKa = 3.12EE483 pKa = 4.32HH484 pKa = 9.22IIFLVDD490 pKa = 3.15GEE492 pKa = 4.59VFHH495 pKa = 6.89TYY497 pKa = 10.32EE498 pKa = 4.69NNPDD502 pKa = 3.39SPFNKK507 pKa = 10.16DD508 pKa = 3.31FFLILNVAMGGTFGGEE524 pKa = 3.72INPDD528 pKa = 3.47FVEE531 pKa = 4.47SSMEE535 pKa = 3.39VDD537 pKa = 4.17YY538 pKa = 11.42IRR540 pKa = 11.84VFQQ543 pKa = 4.13
Molecular weight: 59.62 kDa
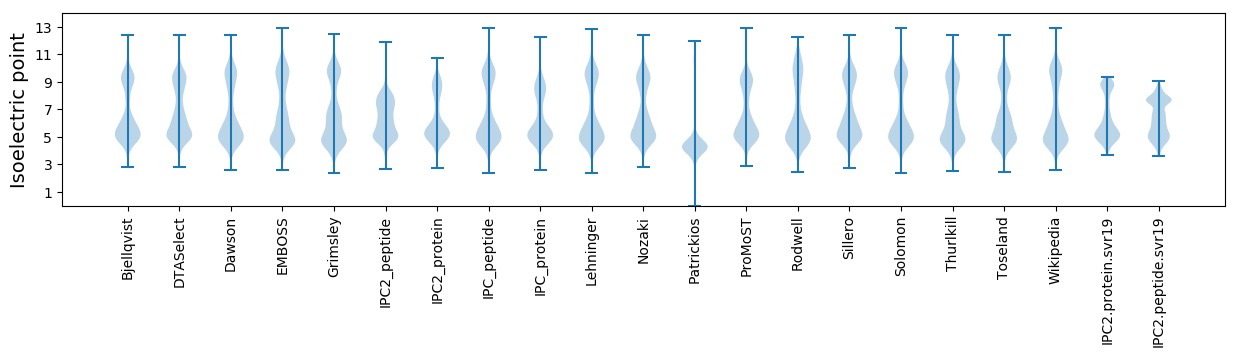
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I4YP95|A0A1I4YP95_9FLAO Ribonucleoside-diphosphate reductase alpha chain OS=Salegentibacter flavus OX=287099 GN=SAMN05660413_00884 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
Molecular weight: 6.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1093900 |
40 |
3354 |
329.6 |
37.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.486 ± 0.033 | 0.642 ± 0.013 |
5.527 ± 0.033 | 7.858 ± 0.052 |
5.176 ± 0.032 | 6.575 ± 0.037 |
1.786 ± 0.021 | 7.623 ± 0.038 |
7.502 ± 0.057 | 9.594 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.29 ± 0.021 | 5.532 ± 0.035 |
3.6 ± 0.021 | 3.418 ± 0.025 |
4.01 ± 0.03 | 6.292 ± 0.032 |
5.099 ± 0.035 | 6.0 ± 0.031 |
1.067 ± 0.017 | 3.923 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
