
Aphanomyces euteiches
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Saprolegniales; Saprolegniaceae; Aphanomyces
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
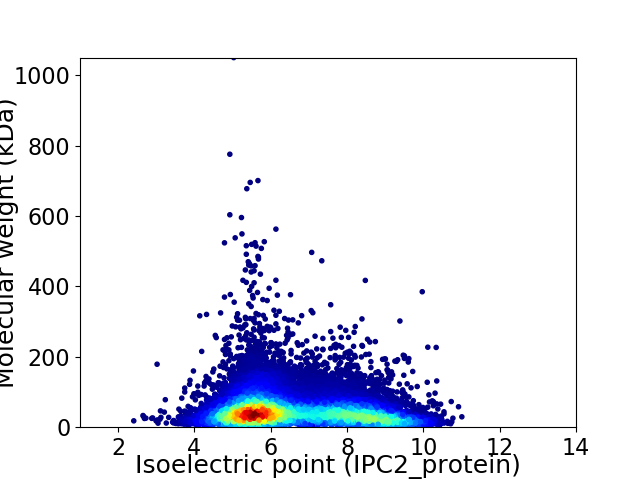
Virtual 2D-PAGE plot for 20021 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
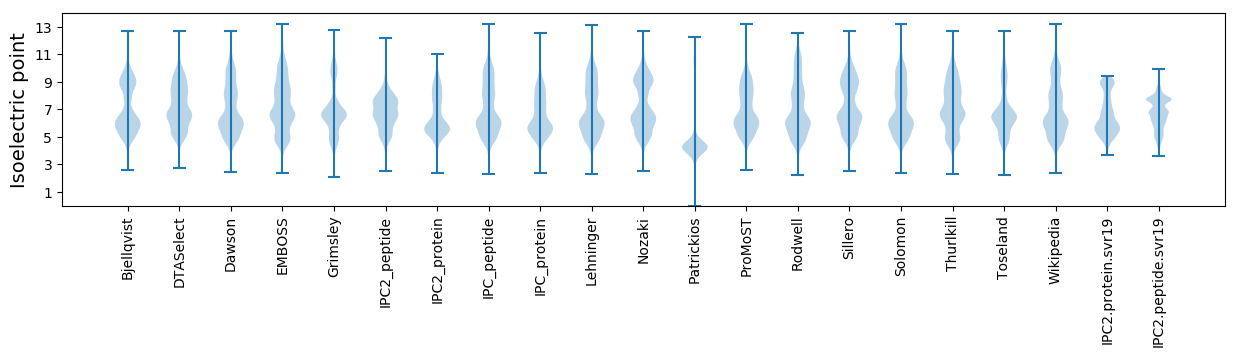
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G0XMT3|A0A6G0XMT3_9STRA Ubiquitin-like domain-containing protein OS=Aphanomyces euteiches OX=100861 GN=Ae201684_003290 PE=4 SV=1
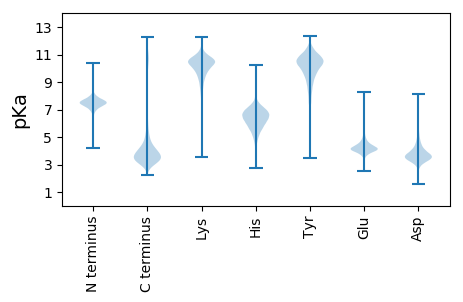
MM1 pKa = 7.3NALHH5 pKa = 6.99LADD8 pKa = 4.71CASNGAPVLIDD19 pKa = 4.11DD20 pKa = 4.69EE21 pKa = 5.18EE22 pKa = 4.74ILASFTNVRR31 pKa = 11.84MFTNTYY37 pKa = 10.55GPGEE41 pKa = 4.59DD42 pKa = 4.9DD43 pKa = 4.81SPVMDD48 pKa = 4.72AKK50 pKa = 11.15GSGNLYY56 pKa = 8.12VTTSRR61 pKa = 11.84VVWVFSQNGSGVVGYY76 pKa = 10.05AWDD79 pKa = 3.51MTYY82 pKa = 11.29LSLHH86 pKa = 6.72AISRR90 pKa = 11.84DD91 pKa = 3.08LSSFPEE97 pKa = 4.17PCLYY101 pKa = 10.58CQLDD105 pKa = 3.64VEE107 pKa = 5.1DD108 pKa = 4.64EE109 pKa = 4.29VNEE112 pKa = 4.0IRR114 pKa = 11.84FVPQDD119 pKa = 3.68PEE121 pKa = 4.04KK122 pKa = 10.6QLQALFDD129 pKa = 4.23AFSEE133 pKa = 4.69SAALNPDD140 pKa = 3.65DD141 pKa = 6.66DD142 pKa = 5.61EE143 pKa = 7.9DD144 pKa = 6.04DD145 pKa = 3.99EE146 pKa = 4.57QQGGDD151 pKa = 3.16WIYY154 pKa = 11.4NEE156 pKa = 4.74EE157 pKa = 4.18EE158 pKa = 4.24VSTGARR164 pKa = 11.84EE165 pKa = 4.0AALAAHH171 pKa = 7.19FDD173 pKa = 4.22SILQVAPSLQQHH185 pKa = 7.0ADD187 pKa = 3.46GQFDD191 pKa = 4.45DD192 pKa = 4.89ADD194 pKa = 4.02EE195 pKa = 6.53DD196 pKa = 3.95EE197 pKa = 4.56MLL199 pKa = 4.82
MM1 pKa = 7.3NALHH5 pKa = 6.99LADD8 pKa = 4.71CASNGAPVLIDD19 pKa = 4.11DD20 pKa = 4.69EE21 pKa = 5.18EE22 pKa = 4.74ILASFTNVRR31 pKa = 11.84MFTNTYY37 pKa = 10.55GPGEE41 pKa = 4.59DD42 pKa = 4.9DD43 pKa = 4.81SPVMDD48 pKa = 4.72AKK50 pKa = 11.15GSGNLYY56 pKa = 8.12VTTSRR61 pKa = 11.84VVWVFSQNGSGVVGYY76 pKa = 10.05AWDD79 pKa = 3.51MTYY82 pKa = 11.29LSLHH86 pKa = 6.72AISRR90 pKa = 11.84DD91 pKa = 3.08LSSFPEE97 pKa = 4.17PCLYY101 pKa = 10.58CQLDD105 pKa = 3.64VEE107 pKa = 5.1DD108 pKa = 4.64EE109 pKa = 4.29VNEE112 pKa = 4.0IRR114 pKa = 11.84FVPQDD119 pKa = 3.68PEE121 pKa = 4.04KK122 pKa = 10.6QLQALFDD129 pKa = 4.23AFSEE133 pKa = 4.69SAALNPDD140 pKa = 3.65DD141 pKa = 6.66DD142 pKa = 5.61EE143 pKa = 7.9DD144 pKa = 6.04DD145 pKa = 3.99EE146 pKa = 4.57QQGGDD151 pKa = 3.16WIYY154 pKa = 11.4NEE156 pKa = 4.74EE157 pKa = 4.18EE158 pKa = 4.24VSTGARR164 pKa = 11.84EE165 pKa = 4.0AALAAHH171 pKa = 7.19FDD173 pKa = 4.22SILQVAPSLQQHH185 pKa = 7.0ADD187 pKa = 3.46GQFDD191 pKa = 4.45DD192 pKa = 4.89ADD194 pKa = 4.02EE195 pKa = 6.53DD196 pKa = 3.95EE197 pKa = 4.56MLL199 pKa = 4.82
Molecular weight: 21.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G0W7F6|A0A6G0W7F6_9STRA Uncharacterized protein OS=Aphanomyces euteiches OX=100861 GN=Ae201684_018062 PE=4 SV=1
MM1 pKa = 7.57GFQLAFSNFLLSMRR15 pKa = 11.84VFLQLFLAAAAAALSHH31 pKa = 6.15GQLANQVVSFEE42 pKa = 4.29VSRR45 pKa = 11.84SLSATAGYY53 pKa = 9.42GALDD57 pKa = 3.64KK58 pKa = 10.52TVQTAAIHH66 pKa = 6.54GEE68 pKa = 3.98DD69 pKa = 3.96TEE71 pKa = 4.52TLRR74 pKa = 11.84SLRR77 pKa = 11.84RR78 pKa = 11.84KK79 pKa = 9.38GRR81 pKa = 11.84GRR83 pKa = 11.84RR84 pKa = 11.84GAKK87 pKa = 9.11SGRR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84NRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84SRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84HH102 pKa = 5.27LMTVQSRR109 pKa = 11.84EE110 pKa = 3.84LAATAGGFMNGLKK123 pKa = 10.85SMGRR127 pKa = 11.84KK128 pKa = 8.94ISSAAGKK135 pKa = 9.85VGRR138 pKa = 11.84AASSATGKK146 pKa = 10.41LSRR149 pKa = 11.84ATSQVGQVAGQVGRR163 pKa = 11.84VAGKK167 pKa = 9.67VSRR170 pKa = 11.84TANRR174 pKa = 11.84VGRR177 pKa = 11.84VAGHH181 pKa = 5.91VSQVGGRR188 pKa = 11.84VGRR191 pKa = 11.84VAGNVGRR198 pKa = 11.84VAGQVRR204 pKa = 11.84RR205 pKa = 11.84GAGQVRR211 pKa = 11.84RR212 pKa = 11.84TAGQVGHH219 pKa = 6.24SARR222 pKa = 11.84QAGRR226 pKa = 11.84AGRR229 pKa = 11.84PTGGRR234 pKa = 11.84SSGGRR239 pKa = 11.84ASGRR243 pKa = 11.84PAGGRR248 pKa = 11.84GRR250 pKa = 11.84ASGSRR255 pKa = 11.84ATGGRR260 pKa = 11.84RR261 pKa = 11.84AAGGRR266 pKa = 11.84ASGGRR271 pKa = 11.84PARR274 pKa = 11.84RR275 pKa = 11.84KK276 pKa = 9.88AGRR279 pKa = 11.84ALRR282 pKa = 11.84GVGSNN287 pKa = 3.48
MM1 pKa = 7.57GFQLAFSNFLLSMRR15 pKa = 11.84VFLQLFLAAAAAALSHH31 pKa = 6.15GQLANQVVSFEE42 pKa = 4.29VSRR45 pKa = 11.84SLSATAGYY53 pKa = 9.42GALDD57 pKa = 3.64KK58 pKa = 10.52TVQTAAIHH66 pKa = 6.54GEE68 pKa = 3.98DD69 pKa = 3.96TEE71 pKa = 4.52TLRR74 pKa = 11.84SLRR77 pKa = 11.84RR78 pKa = 11.84KK79 pKa = 9.38GRR81 pKa = 11.84GRR83 pKa = 11.84RR84 pKa = 11.84GAKK87 pKa = 9.11SGRR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84NRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84SRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84HH102 pKa = 5.27LMTVQSRR109 pKa = 11.84EE110 pKa = 3.84LAATAGGFMNGLKK123 pKa = 10.85SMGRR127 pKa = 11.84KK128 pKa = 8.94ISSAAGKK135 pKa = 9.85VGRR138 pKa = 11.84AASSATGKK146 pKa = 10.41LSRR149 pKa = 11.84ATSQVGQVAGQVGRR163 pKa = 11.84VAGKK167 pKa = 9.67VSRR170 pKa = 11.84TANRR174 pKa = 11.84VGRR177 pKa = 11.84VAGHH181 pKa = 5.91VSQVGGRR188 pKa = 11.84VGRR191 pKa = 11.84VAGNVGRR198 pKa = 11.84VAGQVRR204 pKa = 11.84RR205 pKa = 11.84GAGQVRR211 pKa = 11.84RR212 pKa = 11.84TAGQVGHH219 pKa = 6.24SARR222 pKa = 11.84QAGRR226 pKa = 11.84AGRR229 pKa = 11.84PTGGRR234 pKa = 11.84SSGGRR239 pKa = 11.84ASGRR243 pKa = 11.84PAGGRR248 pKa = 11.84GRR250 pKa = 11.84ASGSRR255 pKa = 11.84ATGGRR260 pKa = 11.84RR261 pKa = 11.84AAGGRR266 pKa = 11.84ASGGRR271 pKa = 11.84PARR274 pKa = 11.84RR275 pKa = 11.84KK276 pKa = 9.88AGRR279 pKa = 11.84ALRR282 pKa = 11.84GVGSNN287 pKa = 3.48
Molecular weight: 29.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
9106444 |
33 |
10036 |
454.8 |
50.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.621 ± 0.02 | 1.794 ± 0.01 |
5.657 ± 0.013 | 5.734 ± 0.019 |
3.964 ± 0.011 | 5.427 ± 0.019 |
2.754 ± 0.009 | 4.811 ± 0.01 |
5.208 ± 0.018 | 9.662 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.536 ± 0.008 | 3.744 ± 0.009 |
5.107 ± 0.017 | 4.289 ± 0.013 |
5.534 ± 0.015 | 7.764 ± 0.019 |
6.164 ± 0.015 | 7.083 ± 0.015 |
1.394 ± 0.006 | 2.748 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
