
Rhizopogon vinicolor AM-OR11-026
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Boletales; Suillineae; Rhizopogonaceae; Rhizopogon; Rhizopogon vinicolor
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
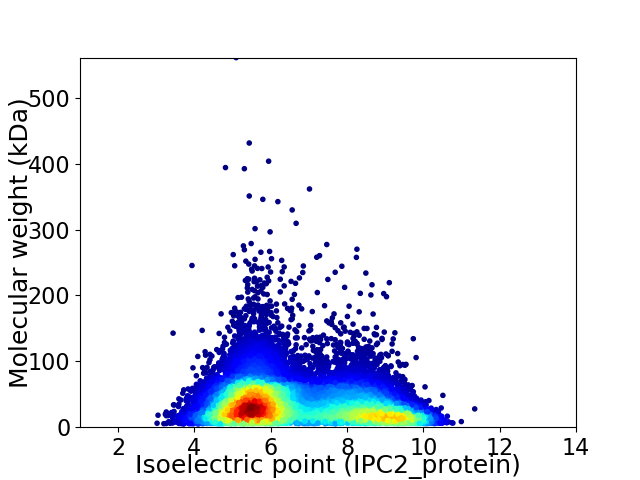
Virtual 2D-PAGE plot for 14447 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B7N717|A0A1B7N717_9AGAM E3 ubiquitin-protein ligase listerin OS=Rhizopogon vinicolor AM-OR11-026 OX=1314800 GN=K503DRAFT_864487 PE=3 SV=1
MM1 pKa = 7.2FNIVNSAFSAVATVFQMIVSYY22 pKa = 9.19MIIALCFIGLMLIDD36 pKa = 5.9LIDD39 pKa = 3.96EE40 pKa = 4.37VSLQVSPASEE50 pKa = 4.12KK51 pKa = 10.54VVCSCVTDD59 pKa = 4.26CPHH62 pKa = 5.8TEE64 pKa = 3.72ARR66 pKa = 11.84EE67 pKa = 3.97EE68 pKa = 4.18LLVAPADD75 pKa = 3.55IDD77 pKa = 3.8EE78 pKa = 4.56VEE80 pKa = 3.84LGRR83 pKa = 11.84RR84 pKa = 11.84YY85 pKa = 10.31VEE87 pKa = 4.14STDD90 pKa = 3.83DD91 pKa = 3.77SDD93 pKa = 3.74VSRR96 pKa = 11.84SFEE99 pKa = 4.0FDD101 pKa = 2.71VSTFSTFEE109 pKa = 4.15SLLSSSPSFIEE120 pKa = 4.88LGSAPWAEE128 pKa = 4.87LDD130 pKa = 3.5TLEE133 pKa = 5.08CVTSSPHH140 pKa = 6.72SPTTTMDD147 pKa = 3.35AADD150 pKa = 3.43ISRR153 pKa = 11.84SFEE156 pKa = 3.83LDD158 pKa = 2.88ASVVGTFEE166 pKa = 4.58FLLSSSPSFIEE177 pKa = 5.36LGSAPCDD184 pKa = 3.42EE185 pKa = 5.72LDD187 pKa = 3.55TPRR190 pKa = 11.84CVTSWMDD197 pKa = 3.03GADD200 pKa = 3.06ISRR203 pKa = 11.84SFEE206 pKa = 4.2LNASVVGTFEE216 pKa = 4.58FLLSSSPSFIEE227 pKa = 5.36LGSAPCDD234 pKa = 3.42EE235 pKa = 5.72LDD237 pKa = 3.61TPRR240 pKa = 11.84CVAFPMDD247 pKa = 4.45DD248 pKa = 3.64ANISRR253 pKa = 11.84SFEE256 pKa = 4.38LNASMVGTFEE266 pKa = 4.67SLLSSSPSFIEE277 pKa = 4.67LGSVPCDD284 pKa = 3.4EE285 pKa = 6.02LDD287 pKa = 3.64TPQCVASSVDD297 pKa = 3.89DD298 pKa = 4.35ADD300 pKa = 3.65ISQSFEE306 pKa = 4.09LDD308 pKa = 3.22ASSLGAFEE316 pKa = 5.12SLLPSSPSFLEE327 pKa = 4.25LGSAPSVEE335 pKa = 4.96LDD337 pKa = 3.38TPEE340 pKa = 4.69CVSSSPYY347 pKa = 10.85LPTTTTMDD355 pKa = 4.08DD356 pKa = 3.57NTSVSLSMFDD366 pKa = 3.24VDD368 pKa = 5.69RR369 pKa = 11.84FPTLISSSTSFVALGEE385 pKa = 4.3ASCSYY390 pKa = 11.19YY391 pKa = 10.77LDD393 pKa = 3.88TSHH396 pKa = 7.01HH397 pKa = 5.58VTSSAPSPNSSVSLSVFDD415 pKa = 5.03LEE417 pKa = 4.93RR418 pKa = 11.84FPSLICPSSSFRR430 pKa = 11.84ALASASRR437 pKa = 11.84RR438 pKa = 11.84SSTASSFSLSTSINDD453 pKa = 3.57ADD455 pKa = 3.98VTISLSSSFSKK466 pKa = 10.98YY467 pKa = 9.89PSLLSSSPSFIALGSVLSS485 pKa = 3.92
MM1 pKa = 7.2FNIVNSAFSAVATVFQMIVSYY22 pKa = 9.19MIIALCFIGLMLIDD36 pKa = 5.9LIDD39 pKa = 3.96EE40 pKa = 4.37VSLQVSPASEE50 pKa = 4.12KK51 pKa = 10.54VVCSCVTDD59 pKa = 4.26CPHH62 pKa = 5.8TEE64 pKa = 3.72ARR66 pKa = 11.84EE67 pKa = 3.97EE68 pKa = 4.18LLVAPADD75 pKa = 3.55IDD77 pKa = 3.8EE78 pKa = 4.56VEE80 pKa = 3.84LGRR83 pKa = 11.84RR84 pKa = 11.84YY85 pKa = 10.31VEE87 pKa = 4.14STDD90 pKa = 3.83DD91 pKa = 3.77SDD93 pKa = 3.74VSRR96 pKa = 11.84SFEE99 pKa = 4.0FDD101 pKa = 2.71VSTFSTFEE109 pKa = 4.15SLLSSSPSFIEE120 pKa = 4.88LGSAPWAEE128 pKa = 4.87LDD130 pKa = 3.5TLEE133 pKa = 5.08CVTSSPHH140 pKa = 6.72SPTTTMDD147 pKa = 3.35AADD150 pKa = 3.43ISRR153 pKa = 11.84SFEE156 pKa = 3.83LDD158 pKa = 2.88ASVVGTFEE166 pKa = 4.58FLLSSSPSFIEE177 pKa = 5.36LGSAPCDD184 pKa = 3.42EE185 pKa = 5.72LDD187 pKa = 3.55TPRR190 pKa = 11.84CVTSWMDD197 pKa = 3.03GADD200 pKa = 3.06ISRR203 pKa = 11.84SFEE206 pKa = 4.2LNASVVGTFEE216 pKa = 4.58FLLSSSPSFIEE227 pKa = 5.36LGSAPCDD234 pKa = 3.42EE235 pKa = 5.72LDD237 pKa = 3.61TPRR240 pKa = 11.84CVAFPMDD247 pKa = 4.45DD248 pKa = 3.64ANISRR253 pKa = 11.84SFEE256 pKa = 4.38LNASMVGTFEE266 pKa = 4.67SLLSSSPSFIEE277 pKa = 4.67LGSVPCDD284 pKa = 3.4EE285 pKa = 6.02LDD287 pKa = 3.64TPQCVASSVDD297 pKa = 3.89DD298 pKa = 4.35ADD300 pKa = 3.65ISQSFEE306 pKa = 4.09LDD308 pKa = 3.22ASSLGAFEE316 pKa = 5.12SLLPSSPSFLEE327 pKa = 4.25LGSAPSVEE335 pKa = 4.96LDD337 pKa = 3.38TPEE340 pKa = 4.69CVSSSPYY347 pKa = 10.85LPTTTTMDD355 pKa = 4.08DD356 pKa = 3.57NTSVSLSMFDD366 pKa = 3.24VDD368 pKa = 5.69RR369 pKa = 11.84FPTLISSSTSFVALGEE385 pKa = 4.3ASCSYY390 pKa = 11.19YY391 pKa = 10.77LDD393 pKa = 3.88TSHH396 pKa = 7.01HH397 pKa = 5.58VTSSAPSPNSSVSLSVFDD415 pKa = 5.03LEE417 pKa = 4.93RR418 pKa = 11.84FPSLICPSSSFRR430 pKa = 11.84ALASASRR437 pKa = 11.84RR438 pKa = 11.84SSTASSFSLSTSINDD453 pKa = 3.57ADD455 pKa = 3.98VTISLSSSFSKK466 pKa = 10.98YY467 pKa = 9.89PSLLSSSPSFIALGSVLSS485 pKa = 3.92
Molecular weight: 51.58 kDa
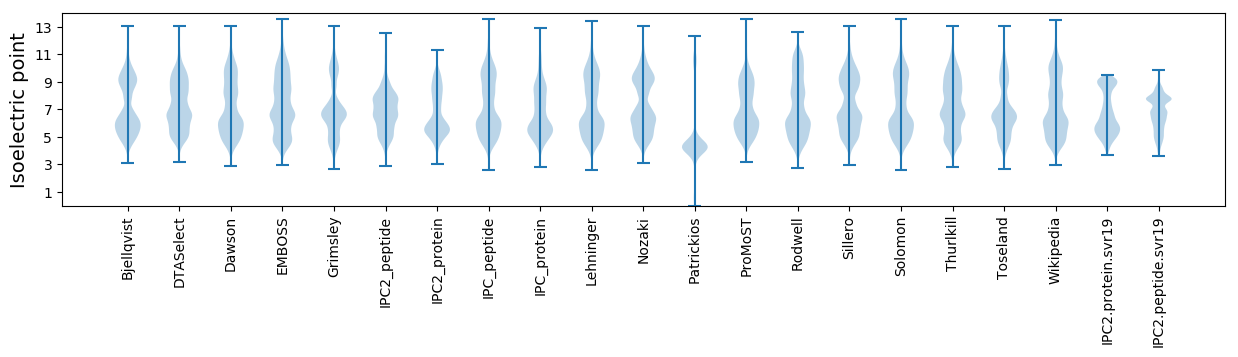
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B7N1U5|A0A1B7N1U5_9AGAM Uncharacterized protein OS=Rhizopogon vinicolor AM-OR11-026 OX=1314800 GN=K503DRAFT_815831 PE=4 SV=1
MM1 pKa = 7.34TRR3 pKa = 11.84TGSPPSRR10 pKa = 11.84SLMILIAMMIKK21 pKa = 10.32SRR23 pKa = 11.84MTVDD27 pKa = 3.53TSHH30 pKa = 7.16TSPTRR35 pKa = 11.84TTTRR39 pKa = 11.84RR40 pKa = 11.84IRR42 pKa = 11.84RR43 pKa = 11.84MIIRR47 pKa = 11.84KK48 pKa = 7.05TSIRR52 pKa = 11.84RR53 pKa = 11.84TIKK56 pKa = 9.94KK57 pKa = 8.27RR58 pKa = 11.84TMISGTRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84TTKK70 pKa = 10.17RR71 pKa = 11.84RR72 pKa = 11.84TKK74 pKa = 10.06RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84TITRR81 pKa = 11.84STMMNGMTTRR91 pKa = 11.84TMTRR95 pKa = 11.84RR96 pKa = 11.84MTRR99 pKa = 11.84RR100 pKa = 11.84MTRR103 pKa = 11.84RR104 pKa = 11.84XXXXTRR110 pKa = 11.84RR111 pKa = 11.84MTRR114 pKa = 11.84RR115 pKa = 11.84MTRR118 pKa = 11.84RR119 pKa = 11.84TTRR122 pKa = 11.84RR123 pKa = 11.84TTRR126 pKa = 11.84KK127 pKa = 6.45ITRR130 pKa = 11.84RR131 pKa = 11.84MTRR134 pKa = 11.84RR135 pKa = 11.84TMMNGTMTRR144 pKa = 11.84RR145 pKa = 11.84MMNGTTTRR153 pKa = 11.84RR154 pKa = 11.84TKK156 pKa = 9.05MRR158 pKa = 11.84KK159 pKa = 8.44IRR161 pKa = 11.84RR162 pKa = 11.84TMTRR166 pKa = 11.84TKK168 pKa = 9.84MIRR171 pKa = 11.84RR172 pKa = 11.84TIMNGTTTTRR182 pKa = 11.84TRR184 pKa = 11.84TRR186 pKa = 11.84RR187 pKa = 11.84TRR189 pKa = 11.84RR190 pKa = 11.84TKK192 pKa = 10.4RR193 pKa = 11.84RR194 pKa = 11.84MMINGTRR201 pKa = 11.84IKK203 pKa = 10.71SGTSLTATLLSNLRR217 pKa = 11.84VIDD220 pKa = 4.13LPKK223 pKa = 10.88KK224 pKa = 10.59NFTCVISYY232 pKa = 7.51TLL234 pKa = 3.54
MM1 pKa = 7.34TRR3 pKa = 11.84TGSPPSRR10 pKa = 11.84SLMILIAMMIKK21 pKa = 10.32SRR23 pKa = 11.84MTVDD27 pKa = 3.53TSHH30 pKa = 7.16TSPTRR35 pKa = 11.84TTTRR39 pKa = 11.84RR40 pKa = 11.84IRR42 pKa = 11.84RR43 pKa = 11.84MIIRR47 pKa = 11.84KK48 pKa = 7.05TSIRR52 pKa = 11.84RR53 pKa = 11.84TIKK56 pKa = 9.94KK57 pKa = 8.27RR58 pKa = 11.84TMISGTRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84TTKK70 pKa = 10.17RR71 pKa = 11.84RR72 pKa = 11.84TKK74 pKa = 10.06RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84TITRR81 pKa = 11.84STMMNGMTTRR91 pKa = 11.84TMTRR95 pKa = 11.84RR96 pKa = 11.84MTRR99 pKa = 11.84RR100 pKa = 11.84MTRR103 pKa = 11.84RR104 pKa = 11.84XXXXTRR110 pKa = 11.84RR111 pKa = 11.84MTRR114 pKa = 11.84RR115 pKa = 11.84MTRR118 pKa = 11.84RR119 pKa = 11.84TTRR122 pKa = 11.84RR123 pKa = 11.84TTRR126 pKa = 11.84KK127 pKa = 6.45ITRR130 pKa = 11.84RR131 pKa = 11.84MTRR134 pKa = 11.84RR135 pKa = 11.84TMMNGTMTRR144 pKa = 11.84RR145 pKa = 11.84MMNGTTTRR153 pKa = 11.84RR154 pKa = 11.84TKK156 pKa = 9.05MRR158 pKa = 11.84KK159 pKa = 8.44IRR161 pKa = 11.84RR162 pKa = 11.84TMTRR166 pKa = 11.84TKK168 pKa = 9.84MIRR171 pKa = 11.84RR172 pKa = 11.84TIMNGTTTTRR182 pKa = 11.84TRR184 pKa = 11.84TRR186 pKa = 11.84RR187 pKa = 11.84TRR189 pKa = 11.84RR190 pKa = 11.84TKK192 pKa = 10.4RR193 pKa = 11.84RR194 pKa = 11.84MMINGTRR201 pKa = 11.84IKK203 pKa = 10.71SGTSLTATLLSNLRR217 pKa = 11.84VIDD220 pKa = 4.13LPKK223 pKa = 10.88KK224 pKa = 10.59NFTCVISYY232 pKa = 7.51TLL234 pKa = 3.54
Molecular weight: 27.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5225860 |
49 |
5049 |
361.7 |
40.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.113 ± 0.019 | 1.442 ± 0.01 |
5.545 ± 0.015 | 5.643 ± 0.022 |
3.822 ± 0.014 | 6.25 ± 0.021 |
2.721 ± 0.011 | 5.241 ± 0.016 |
4.371 ± 0.019 | 9.383 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.231 ± 0.008 | 3.409 ± 0.011 |
6.239 ± 0.029 | 3.753 ± 0.011 |
6.134 ± 0.019 | 8.941 ± 0.03 |
6.073 ± 0.014 | 6.469 ± 0.016 |
1.492 ± 0.009 | 2.723 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
