
Coniothyrium minitans RNA virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
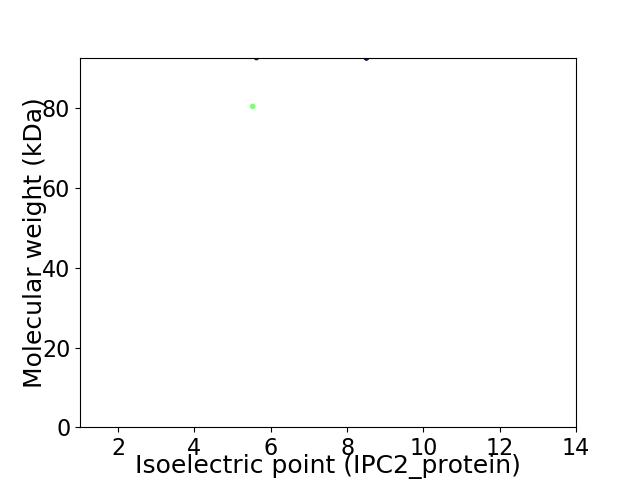
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8B5B7|Q8B5B7_9VIRU RNA-directed RNA polymerase OS=Coniothyrium minitans RNA virus OX=210827 PE=3 SV=1
MM1 pKa = 7.53ASSTRR6 pKa = 11.84FLASAASPLTGTVAGISAGTIGQDD30 pKa = 3.04NQYY33 pKa = 10.34RR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 9.92RR37 pKa = 11.84AGLTIGVHH45 pKa = 4.54EE46 pKa = 5.09HH47 pKa = 6.62GSYY50 pKa = 8.47TQARR54 pKa = 11.84RR55 pKa = 11.84SIFYY59 pKa = 10.28EE60 pKa = 3.51VGRR63 pKa = 11.84RR64 pKa = 11.84YY65 pKa = 10.5GRR67 pKa = 11.84LTDD70 pKa = 3.53ALGPHH75 pKa = 6.49GAEE78 pKa = 4.25AVPIDD83 pKa = 3.43ASVRR87 pKa = 11.84INAAEE92 pKa = 3.86AANFEE97 pKa = 4.0GFARR101 pKa = 11.84KK102 pKa = 9.39FSNFSPQWLTMDD114 pKa = 4.02LCGIAEE120 pKa = 4.11RR121 pKa = 11.84LAKK124 pKa = 10.15GVAAQSVYY132 pKa = 11.16GGVNIVNLRR141 pKa = 11.84GHH143 pKa = 6.39LPVRR147 pKa = 11.84VVALGTLDD155 pKa = 4.0SPQTASNNSVFIPRR169 pKa = 11.84TVDD172 pKa = 3.11TVGNDD177 pKa = 2.94HH178 pKa = 6.29VFAVLAAAANGEE190 pKa = 4.2GAAVSTDD197 pKa = 3.53VLRR200 pKa = 11.84LDD202 pKa = 4.43ANTNEE207 pKa = 4.11PVIPAVSGPSLASACIEE224 pKa = 3.77ALRR227 pKa = 11.84IVGANMEE234 pKa = 4.26ASGAGDD240 pKa = 3.99LFAYY244 pKa = 10.12AVTRR248 pKa = 11.84GIHH251 pKa = 5.54AVVSVVAHH259 pKa = 5.89TDD261 pKa = 2.67EE262 pKa = 4.79GGYY265 pKa = 8.74MRR267 pKa = 11.84ALLRR271 pKa = 11.84HH272 pKa = 4.93GRR274 pKa = 11.84FRR276 pKa = 11.84VPYY279 pKa = 10.14GGINQALRR287 pKa = 11.84DD288 pKa = 4.12YY289 pKa = 9.94PALPAAGALATHH301 pKa = 6.96VISSWVDD308 pKa = 3.93AIALKK313 pKa = 9.09TAAVVAHH320 pKa = 6.79SDD322 pKa = 3.37PCVIASGGLYY332 pKa = 7.65PTVFTSSQGDD342 pKa = 3.42ITPPGTDD349 pKa = 3.65EE350 pKa = 4.94GDD352 pKa = 3.69SPTDD356 pKa = 3.2ADD358 pKa = 3.3ARR360 pKa = 11.84AIGRR364 pKa = 11.84QIAGDD369 pKa = 4.26LGRR372 pKa = 11.84FAPTFMAGLLRR383 pKa = 11.84IFGLQTSSQVAEE395 pKa = 3.83AHH397 pKa = 5.93FCTVGGMYY405 pKa = 10.6LSEE408 pKa = 4.15NVDD411 pKa = 2.59RR412 pKa = 11.84HH413 pKa = 5.7LRR415 pKa = 11.84HH416 pKa = 5.82KK417 pKa = 9.35TVAPYY422 pKa = 9.94FWVEE426 pKa = 3.75PTSLIEE432 pKa = 4.04VGFLGSTAEE441 pKa = 4.12TAGFGSLVTPGDD453 pKa = 3.21QAMIPTFEE461 pKa = 4.75RR462 pKa = 11.84VRR464 pKa = 11.84EE465 pKa = 3.88MDD467 pKa = 2.88RR468 pKa = 11.84GRR470 pKa = 11.84NANFSTIAFKK480 pKa = 10.48MRR482 pKa = 11.84TARR485 pKa = 11.84TSGLVCAHH493 pKa = 7.18AAAPTPLAGLKK504 pKa = 10.11LYY506 pKa = 10.81QFDD509 pKa = 3.83QDD511 pKa = 4.16SLILAGDD518 pKa = 3.59QGPTNGDD525 pKa = 3.27VPTKK529 pKa = 10.42HH530 pKa = 5.96VAADD534 pKa = 3.83PLSSYY539 pKa = 10.76LWVRR543 pKa = 11.84GQSAIPAPAEE553 pKa = 4.06MINTQASYY561 pKa = 9.88AAKK564 pKa = 9.99YY565 pKa = 10.59KK566 pKa = 10.86NITWDD571 pKa = 3.54DD572 pKa = 3.84DD573 pKa = 4.34FEE575 pKa = 4.69GTVSDD580 pKa = 5.69LPKK583 pKa = 10.44AWEE586 pKa = 4.33LEE588 pKa = 4.02HH589 pKa = 6.6DD590 pKa = 3.99TMWRR594 pKa = 11.84VTVPTAFSVTGPSNYY609 pKa = 9.69LDD611 pKa = 3.53RR612 pKa = 11.84EE613 pKa = 4.09ARR615 pKa = 11.84RR616 pKa = 11.84ARR618 pKa = 11.84SRR620 pKa = 11.84AAISLAQAALRR631 pKa = 11.84ARR633 pKa = 11.84AYY635 pKa = 10.5GEE637 pKa = 4.19ANSPVIDD644 pKa = 4.03VSNVPPTWDD653 pKa = 3.5DD654 pKa = 2.86EE655 pKa = 4.37RR656 pKa = 11.84APVVVFSDD664 pKa = 4.98EE665 pKa = 3.89IHH667 pKa = 6.27TSDD670 pKa = 4.47PGTAQGRR677 pKa = 11.84GIGNPPDD684 pKa = 3.15VTAPVTGAPLVPIPHH699 pKa = 6.42HH700 pKa = 5.9QPLRR704 pKa = 11.84GPPFPRR710 pKa = 11.84GAGVMGGGVAPPPPAGPGGPPAGPGPNPPPPPPPGGDD747 pKa = 3.36GGDD750 pKa = 3.8DD751 pKa = 3.26AAAAAAVAGGDD762 pKa = 3.43VHH764 pKa = 6.68GAAGALPAPQVV775 pKa = 3.24
MM1 pKa = 7.53ASSTRR6 pKa = 11.84FLASAASPLTGTVAGISAGTIGQDD30 pKa = 3.04NQYY33 pKa = 10.34RR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 9.92RR37 pKa = 11.84AGLTIGVHH45 pKa = 4.54EE46 pKa = 5.09HH47 pKa = 6.62GSYY50 pKa = 8.47TQARR54 pKa = 11.84RR55 pKa = 11.84SIFYY59 pKa = 10.28EE60 pKa = 3.51VGRR63 pKa = 11.84RR64 pKa = 11.84YY65 pKa = 10.5GRR67 pKa = 11.84LTDD70 pKa = 3.53ALGPHH75 pKa = 6.49GAEE78 pKa = 4.25AVPIDD83 pKa = 3.43ASVRR87 pKa = 11.84INAAEE92 pKa = 3.86AANFEE97 pKa = 4.0GFARR101 pKa = 11.84KK102 pKa = 9.39FSNFSPQWLTMDD114 pKa = 4.02LCGIAEE120 pKa = 4.11RR121 pKa = 11.84LAKK124 pKa = 10.15GVAAQSVYY132 pKa = 11.16GGVNIVNLRR141 pKa = 11.84GHH143 pKa = 6.39LPVRR147 pKa = 11.84VVALGTLDD155 pKa = 4.0SPQTASNNSVFIPRR169 pKa = 11.84TVDD172 pKa = 3.11TVGNDD177 pKa = 2.94HH178 pKa = 6.29VFAVLAAAANGEE190 pKa = 4.2GAAVSTDD197 pKa = 3.53VLRR200 pKa = 11.84LDD202 pKa = 4.43ANTNEE207 pKa = 4.11PVIPAVSGPSLASACIEE224 pKa = 3.77ALRR227 pKa = 11.84IVGANMEE234 pKa = 4.26ASGAGDD240 pKa = 3.99LFAYY244 pKa = 10.12AVTRR248 pKa = 11.84GIHH251 pKa = 5.54AVVSVVAHH259 pKa = 5.89TDD261 pKa = 2.67EE262 pKa = 4.79GGYY265 pKa = 8.74MRR267 pKa = 11.84ALLRR271 pKa = 11.84HH272 pKa = 4.93GRR274 pKa = 11.84FRR276 pKa = 11.84VPYY279 pKa = 10.14GGINQALRR287 pKa = 11.84DD288 pKa = 4.12YY289 pKa = 9.94PALPAAGALATHH301 pKa = 6.96VISSWVDD308 pKa = 3.93AIALKK313 pKa = 9.09TAAVVAHH320 pKa = 6.79SDD322 pKa = 3.37PCVIASGGLYY332 pKa = 7.65PTVFTSSQGDD342 pKa = 3.42ITPPGTDD349 pKa = 3.65EE350 pKa = 4.94GDD352 pKa = 3.69SPTDD356 pKa = 3.2ADD358 pKa = 3.3ARR360 pKa = 11.84AIGRR364 pKa = 11.84QIAGDD369 pKa = 4.26LGRR372 pKa = 11.84FAPTFMAGLLRR383 pKa = 11.84IFGLQTSSQVAEE395 pKa = 3.83AHH397 pKa = 5.93FCTVGGMYY405 pKa = 10.6LSEE408 pKa = 4.15NVDD411 pKa = 2.59RR412 pKa = 11.84HH413 pKa = 5.7LRR415 pKa = 11.84HH416 pKa = 5.82KK417 pKa = 9.35TVAPYY422 pKa = 9.94FWVEE426 pKa = 3.75PTSLIEE432 pKa = 4.04VGFLGSTAEE441 pKa = 4.12TAGFGSLVTPGDD453 pKa = 3.21QAMIPTFEE461 pKa = 4.75RR462 pKa = 11.84VRR464 pKa = 11.84EE465 pKa = 3.88MDD467 pKa = 2.88RR468 pKa = 11.84GRR470 pKa = 11.84NANFSTIAFKK480 pKa = 10.48MRR482 pKa = 11.84TARR485 pKa = 11.84TSGLVCAHH493 pKa = 7.18AAAPTPLAGLKK504 pKa = 10.11LYY506 pKa = 10.81QFDD509 pKa = 3.83QDD511 pKa = 4.16SLILAGDD518 pKa = 3.59QGPTNGDD525 pKa = 3.27VPTKK529 pKa = 10.42HH530 pKa = 5.96VAADD534 pKa = 3.83PLSSYY539 pKa = 10.76LWVRR543 pKa = 11.84GQSAIPAPAEE553 pKa = 4.06MINTQASYY561 pKa = 9.88AAKK564 pKa = 9.99YY565 pKa = 10.59KK566 pKa = 10.86NITWDD571 pKa = 3.54DD572 pKa = 3.84DD573 pKa = 4.34FEE575 pKa = 4.69GTVSDD580 pKa = 5.69LPKK583 pKa = 10.44AWEE586 pKa = 4.33LEE588 pKa = 4.02HH589 pKa = 6.6DD590 pKa = 3.99TMWRR594 pKa = 11.84VTVPTAFSVTGPSNYY609 pKa = 9.69LDD611 pKa = 3.53RR612 pKa = 11.84EE613 pKa = 4.09ARR615 pKa = 11.84RR616 pKa = 11.84ARR618 pKa = 11.84SRR620 pKa = 11.84AAISLAQAALRR631 pKa = 11.84ARR633 pKa = 11.84AYY635 pKa = 10.5GEE637 pKa = 4.19ANSPVIDD644 pKa = 4.03VSNVPPTWDD653 pKa = 3.5DD654 pKa = 2.86EE655 pKa = 4.37RR656 pKa = 11.84APVVVFSDD664 pKa = 4.98EE665 pKa = 3.89IHH667 pKa = 6.27TSDD670 pKa = 4.47PGTAQGRR677 pKa = 11.84GIGNPPDD684 pKa = 3.15VTAPVTGAPLVPIPHH699 pKa = 6.42HH700 pKa = 5.9QPLRR704 pKa = 11.84GPPFPRR710 pKa = 11.84GAGVMGGGVAPPPPAGPGGPPAGPGPNPPPPPPPGGDD747 pKa = 3.36GGDD750 pKa = 3.8DD751 pKa = 3.26AAAAAAVAGGDD762 pKa = 3.43VHH764 pKa = 6.68GAAGALPAPQVV775 pKa = 3.24
Molecular weight: 80.52 kDa
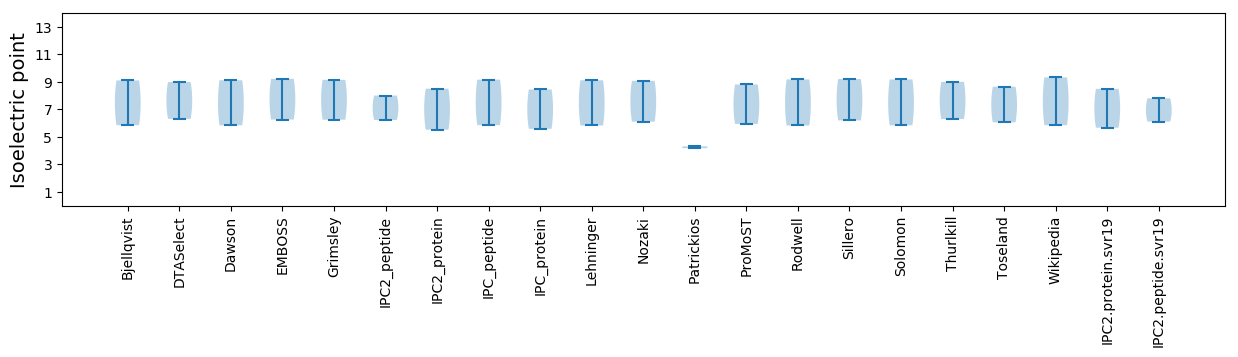
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8B5B7|Q8B5B7_9VIRU RNA-directed RNA polymerase OS=Coniothyrium minitans RNA virus OX=210827 PE=3 SV=1
MM1 pKa = 7.21IGARR5 pKa = 11.84VDD7 pKa = 3.69EE8 pKa = 4.53RR9 pKa = 11.84ALAAGALGQYY19 pKa = 9.45LKK21 pKa = 11.04KK22 pKa = 9.85LTRR25 pKa = 11.84SQDD28 pKa = 3.1VSRR31 pKa = 11.84FATLSFPDD39 pKa = 3.39QISYY43 pKa = 9.97VFKK46 pKa = 10.53PSWDD50 pKa = 3.52SFRR53 pKa = 11.84PTPLHH58 pKa = 6.14RR59 pKa = 11.84AALSFLCGEE68 pKa = 4.48VPVQVALPYY77 pKa = 9.98ASLWRR82 pKa = 11.84MVDD85 pKa = 3.28YY86 pKa = 8.78TCPVPDD92 pKa = 3.26IVLDD96 pKa = 3.85IKK98 pKa = 11.44YY99 pKa = 6.82KK100 pKa = 9.22TRR102 pKa = 11.84WIRR105 pKa = 11.84DD106 pKa = 2.95AGITVKK112 pKa = 10.57RR113 pKa = 11.84FPLKK117 pKa = 10.58KK118 pKa = 10.47NPAASNKK125 pKa = 9.07VNLYY129 pKa = 8.52LHH131 pKa = 6.41EE132 pKa = 4.45VGRR135 pKa = 11.84DD136 pKa = 3.75LSSHH140 pKa = 6.81APTQLNVGLQYY151 pKa = 10.99LDD153 pKa = 2.95WVRR156 pKa = 11.84RR157 pKa = 11.84QRR159 pKa = 11.84VVYY162 pKa = 10.23NDD164 pKa = 2.99QATAFLLYY172 pKa = 9.36EE173 pKa = 4.17YY174 pKa = 10.74CLACYY179 pKa = 10.03DD180 pKa = 3.76SRR182 pKa = 11.84GWRR185 pKa = 11.84HH186 pKa = 5.41AAASLFDD193 pKa = 4.01SDD195 pKa = 4.22YY196 pKa = 11.69AKK198 pKa = 10.82GFSVFGKK205 pKa = 10.33AVGINGSQVGAMMVEE220 pKa = 4.42TNVLLGRR227 pKa = 11.84DD228 pKa = 3.78VAPIDD233 pKa = 3.77LVEE236 pKa = 4.32EE237 pKa = 4.04ARR239 pKa = 11.84KK240 pKa = 7.51RR241 pKa = 11.84TSMRR245 pKa = 11.84EE246 pKa = 3.75VRR248 pKa = 11.84AMTTAYY254 pKa = 10.76SDD256 pKa = 3.35DD257 pKa = 3.93TIRR260 pKa = 11.84RR261 pKa = 11.84AVRR264 pKa = 11.84TVLLRR269 pKa = 11.84EE270 pKa = 4.67LKK272 pKa = 10.55RR273 pKa = 11.84SGDD276 pKa = 3.74SYY278 pKa = 11.3ILEE281 pKa = 4.49FPTLEE286 pKa = 4.18DD287 pKa = 2.61HH288 pKa = 7.29WATRR292 pKa = 11.84WQWAVNGAHH301 pKa = 6.62SGLIYY306 pKa = 9.33KK307 pKa = 7.59TNPSYY312 pKa = 10.55RR313 pKa = 11.84PHH315 pKa = 6.25MPGFDD320 pKa = 3.4RR321 pKa = 11.84LHH323 pKa = 5.75RR324 pKa = 11.84RR325 pKa = 11.84AWLEE329 pKa = 4.08TISEE333 pKa = 4.42DD334 pKa = 5.38PRR336 pKa = 11.84PAWDD340 pKa = 2.94GRR342 pKa = 11.84TYY344 pKa = 11.29VSASPKK350 pKa = 10.08LEE352 pKa = 4.2HH353 pKa = 6.54GKK355 pKa = 8.03TRR357 pKa = 11.84AIFAWDD363 pKa = 4.4TINSLAMEE371 pKa = 4.67HH372 pKa = 6.61LMSTVEE378 pKa = 4.06ANWRR382 pKa = 11.84GEE384 pKa = 3.99RR385 pKa = 11.84VILNPGKK392 pKa = 10.51GGHH395 pKa = 6.87LGMAQRR401 pKa = 11.84VQAARR406 pKa = 11.84NRR408 pKa = 11.84SGVSLMLDD416 pKa = 3.09YY417 pKa = 11.57DD418 pKa = 4.5DD419 pKa = 6.0FNSHH423 pKa = 5.25HH424 pKa = 6.14TTRR427 pKa = 11.84TMQIVVEE434 pKa = 4.38EE435 pKa = 4.39TCRR438 pKa = 11.84LTGYY442 pKa = 9.98PPDD445 pKa = 3.83LAEE448 pKa = 4.58KK449 pKa = 10.14LVSSMEE455 pKa = 3.64KK456 pKa = 10.23HH457 pKa = 5.96YY458 pKa = 10.84ICVGGAYY465 pKa = 9.42IGRR468 pKa = 11.84SKK470 pKa = 9.52GTLMSGHH477 pKa = 7.24RR478 pKa = 11.84LTTYY482 pKa = 10.43INSVCNEE489 pKa = 3.51AYY491 pKa = 10.57LRR493 pKa = 11.84IEE495 pKa = 4.77LGDD498 pKa = 4.31DD499 pKa = 3.65FLDD502 pKa = 4.16KK503 pKa = 11.01NVSLHH508 pKa = 6.41VGDD511 pKa = 5.06DD512 pKa = 3.49VYY514 pKa = 11.55LGVRR518 pKa = 11.84SYY520 pKa = 11.46QEE522 pKa = 3.25AGYY525 pKa = 8.45VLRR528 pKa = 11.84MIRR531 pKa = 11.84GSKK534 pKa = 10.43LRR536 pKa = 11.84MNPAKK541 pKa = 10.37QSVGHH546 pKa = 5.33VTTEE550 pKa = 3.91FLRR553 pKa = 11.84VASEE557 pKa = 3.78SRR559 pKa = 11.84YY560 pKa = 9.91SYY562 pKa = 11.03GYY564 pKa = 9.65LARR567 pKa = 11.84AVASITSGSWVNEE580 pKa = 3.64LALAPLEE587 pKa = 4.13ALTNIVASARR597 pKa = 11.84SLANRR602 pKa = 11.84SGIADD607 pKa = 3.57VALLLVSSTRR617 pKa = 11.84RR618 pKa = 11.84MAPLDD623 pKa = 3.75SRR625 pKa = 11.84DD626 pKa = 3.64DD627 pKa = 3.61TLLRR631 pKa = 11.84EE632 pKa = 4.2LLTGKK637 pKa = 9.89VALQNGPNYY646 pKa = 9.89QSSGYY651 pKa = 7.94YY652 pKa = 9.04RR653 pKa = 11.84HH654 pKa = 6.12VAVTPKK660 pKa = 9.69MVRR663 pKa = 11.84RR664 pKa = 11.84DD665 pKa = 3.52DD666 pKa = 3.62FGYY669 pKa = 11.04GVLPLEE675 pKa = 4.5ATHH678 pKa = 6.98TYY680 pKa = 10.22LSSAATVLEE689 pKa = 4.26IEE691 pKa = 4.59TLTKK695 pKa = 10.67AGISVEE701 pKa = 3.77EE702 pKa = 4.18DD703 pKa = 2.94MARR706 pKa = 11.84ASYY709 pKa = 10.25KK710 pKa = 10.35KK711 pKa = 10.1SAPRR715 pKa = 11.84DD716 pKa = 3.65FFSAEE721 pKa = 4.14CLVTGPVLQRR731 pKa = 11.84PCVGVEE737 pKa = 3.43WAEE740 pKa = 3.95MVLRR744 pKa = 11.84RR745 pKa = 11.84PRR747 pKa = 11.84VTGILSRR754 pKa = 11.84YY755 pKa = 8.67PLLLLARR762 pKa = 11.84YY763 pKa = 8.1RR764 pKa = 11.84LPEE767 pKa = 3.91RR768 pKa = 11.84VVRR771 pKa = 11.84EE772 pKa = 3.94ALAAAGGDD780 pKa = 3.88YY781 pKa = 7.67NTPYY785 pKa = 10.73LDD787 pKa = 3.26YY788 pKa = 10.94DD789 pKa = 3.11AWGEE793 pKa = 4.05YY794 pKa = 10.72AHH796 pKa = 6.88GCVIDD801 pKa = 3.86TVMSYY806 pKa = 10.55TDD808 pKa = 3.85ASALGAKK815 pKa = 8.08TAAGVLTSTTRR826 pKa = 11.84MYY828 pKa = 11.15VV829 pKa = 2.83
MM1 pKa = 7.21IGARR5 pKa = 11.84VDD7 pKa = 3.69EE8 pKa = 4.53RR9 pKa = 11.84ALAAGALGQYY19 pKa = 9.45LKK21 pKa = 11.04KK22 pKa = 9.85LTRR25 pKa = 11.84SQDD28 pKa = 3.1VSRR31 pKa = 11.84FATLSFPDD39 pKa = 3.39QISYY43 pKa = 9.97VFKK46 pKa = 10.53PSWDD50 pKa = 3.52SFRR53 pKa = 11.84PTPLHH58 pKa = 6.14RR59 pKa = 11.84AALSFLCGEE68 pKa = 4.48VPVQVALPYY77 pKa = 9.98ASLWRR82 pKa = 11.84MVDD85 pKa = 3.28YY86 pKa = 8.78TCPVPDD92 pKa = 3.26IVLDD96 pKa = 3.85IKK98 pKa = 11.44YY99 pKa = 6.82KK100 pKa = 9.22TRR102 pKa = 11.84WIRR105 pKa = 11.84DD106 pKa = 2.95AGITVKK112 pKa = 10.57RR113 pKa = 11.84FPLKK117 pKa = 10.58KK118 pKa = 10.47NPAASNKK125 pKa = 9.07VNLYY129 pKa = 8.52LHH131 pKa = 6.41EE132 pKa = 4.45VGRR135 pKa = 11.84DD136 pKa = 3.75LSSHH140 pKa = 6.81APTQLNVGLQYY151 pKa = 10.99LDD153 pKa = 2.95WVRR156 pKa = 11.84RR157 pKa = 11.84QRR159 pKa = 11.84VVYY162 pKa = 10.23NDD164 pKa = 2.99QATAFLLYY172 pKa = 9.36EE173 pKa = 4.17YY174 pKa = 10.74CLACYY179 pKa = 10.03DD180 pKa = 3.76SRR182 pKa = 11.84GWRR185 pKa = 11.84HH186 pKa = 5.41AAASLFDD193 pKa = 4.01SDD195 pKa = 4.22YY196 pKa = 11.69AKK198 pKa = 10.82GFSVFGKK205 pKa = 10.33AVGINGSQVGAMMVEE220 pKa = 4.42TNVLLGRR227 pKa = 11.84DD228 pKa = 3.78VAPIDD233 pKa = 3.77LVEE236 pKa = 4.32EE237 pKa = 4.04ARR239 pKa = 11.84KK240 pKa = 7.51RR241 pKa = 11.84TSMRR245 pKa = 11.84EE246 pKa = 3.75VRR248 pKa = 11.84AMTTAYY254 pKa = 10.76SDD256 pKa = 3.35DD257 pKa = 3.93TIRR260 pKa = 11.84RR261 pKa = 11.84AVRR264 pKa = 11.84TVLLRR269 pKa = 11.84EE270 pKa = 4.67LKK272 pKa = 10.55RR273 pKa = 11.84SGDD276 pKa = 3.74SYY278 pKa = 11.3ILEE281 pKa = 4.49FPTLEE286 pKa = 4.18DD287 pKa = 2.61HH288 pKa = 7.29WATRR292 pKa = 11.84WQWAVNGAHH301 pKa = 6.62SGLIYY306 pKa = 9.33KK307 pKa = 7.59TNPSYY312 pKa = 10.55RR313 pKa = 11.84PHH315 pKa = 6.25MPGFDD320 pKa = 3.4RR321 pKa = 11.84LHH323 pKa = 5.75RR324 pKa = 11.84RR325 pKa = 11.84AWLEE329 pKa = 4.08TISEE333 pKa = 4.42DD334 pKa = 5.38PRR336 pKa = 11.84PAWDD340 pKa = 2.94GRR342 pKa = 11.84TYY344 pKa = 11.29VSASPKK350 pKa = 10.08LEE352 pKa = 4.2HH353 pKa = 6.54GKK355 pKa = 8.03TRR357 pKa = 11.84AIFAWDD363 pKa = 4.4TINSLAMEE371 pKa = 4.67HH372 pKa = 6.61LMSTVEE378 pKa = 4.06ANWRR382 pKa = 11.84GEE384 pKa = 3.99RR385 pKa = 11.84VILNPGKK392 pKa = 10.51GGHH395 pKa = 6.87LGMAQRR401 pKa = 11.84VQAARR406 pKa = 11.84NRR408 pKa = 11.84SGVSLMLDD416 pKa = 3.09YY417 pKa = 11.57DD418 pKa = 4.5DD419 pKa = 6.0FNSHH423 pKa = 5.25HH424 pKa = 6.14TTRR427 pKa = 11.84TMQIVVEE434 pKa = 4.38EE435 pKa = 4.39TCRR438 pKa = 11.84LTGYY442 pKa = 9.98PPDD445 pKa = 3.83LAEE448 pKa = 4.58KK449 pKa = 10.14LVSSMEE455 pKa = 3.64KK456 pKa = 10.23HH457 pKa = 5.96YY458 pKa = 10.84ICVGGAYY465 pKa = 9.42IGRR468 pKa = 11.84SKK470 pKa = 9.52GTLMSGHH477 pKa = 7.24RR478 pKa = 11.84LTTYY482 pKa = 10.43INSVCNEE489 pKa = 3.51AYY491 pKa = 10.57LRR493 pKa = 11.84IEE495 pKa = 4.77LGDD498 pKa = 4.31DD499 pKa = 3.65FLDD502 pKa = 4.16KK503 pKa = 11.01NVSLHH508 pKa = 6.41VGDD511 pKa = 5.06DD512 pKa = 3.49VYY514 pKa = 11.55LGVRR518 pKa = 11.84SYY520 pKa = 11.46QEE522 pKa = 3.25AGYY525 pKa = 8.45VLRR528 pKa = 11.84MIRR531 pKa = 11.84GSKK534 pKa = 10.43LRR536 pKa = 11.84MNPAKK541 pKa = 10.37QSVGHH546 pKa = 5.33VTTEE550 pKa = 3.91FLRR553 pKa = 11.84VASEE557 pKa = 3.78SRR559 pKa = 11.84YY560 pKa = 9.91SYY562 pKa = 11.03GYY564 pKa = 9.65LARR567 pKa = 11.84AVASITSGSWVNEE580 pKa = 3.64LALAPLEE587 pKa = 4.13ALTNIVASARR597 pKa = 11.84SLANRR602 pKa = 11.84SGIADD607 pKa = 3.57VALLLVSSTRR617 pKa = 11.84RR618 pKa = 11.84MAPLDD623 pKa = 3.75SRR625 pKa = 11.84DD626 pKa = 3.64DD627 pKa = 3.61TLLRR631 pKa = 11.84EE632 pKa = 4.2LLTGKK637 pKa = 9.89VALQNGPNYY646 pKa = 9.89QSSGYY651 pKa = 7.94YY652 pKa = 9.04RR653 pKa = 11.84HH654 pKa = 6.12VAVTPKK660 pKa = 9.69MVRR663 pKa = 11.84RR664 pKa = 11.84DD665 pKa = 3.52DD666 pKa = 3.62FGYY669 pKa = 11.04GVLPLEE675 pKa = 4.5ATHH678 pKa = 6.98TYY680 pKa = 10.22LSSAATVLEE689 pKa = 4.26IEE691 pKa = 4.59TLTKK695 pKa = 10.67AGISVEE701 pKa = 3.77EE702 pKa = 4.18DD703 pKa = 2.94MARR706 pKa = 11.84ASYY709 pKa = 10.25KK710 pKa = 10.35KK711 pKa = 10.1SAPRR715 pKa = 11.84DD716 pKa = 3.65FFSAEE721 pKa = 4.14CLVTGPVLQRR731 pKa = 11.84PCVGVEE737 pKa = 3.43WAEE740 pKa = 3.95MVLRR744 pKa = 11.84RR745 pKa = 11.84PRR747 pKa = 11.84VTGILSRR754 pKa = 11.84YY755 pKa = 8.67PLLLLARR762 pKa = 11.84YY763 pKa = 8.1RR764 pKa = 11.84LPEE767 pKa = 3.91RR768 pKa = 11.84VVRR771 pKa = 11.84EE772 pKa = 3.94ALAAAGGDD780 pKa = 3.88YY781 pKa = 7.67NTPYY785 pKa = 10.73LDD787 pKa = 3.26YY788 pKa = 10.94DD789 pKa = 3.11AWGEE793 pKa = 4.05YY794 pKa = 10.72AHH796 pKa = 6.88GCVIDD801 pKa = 3.86TVMSYY806 pKa = 10.55TDD808 pKa = 3.85ASALGAKK815 pKa = 8.08TAAGVLTSTTRR826 pKa = 11.84MYY828 pKa = 11.15VV829 pKa = 2.83
Molecular weight: 92.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1604 |
775 |
829 |
802.0 |
86.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.282 ± 1.689 | 0.935 ± 0.192 |
5.611 ± 0.044 | 4.177 ± 0.458 |
2.805 ± 0.363 | 8.666 ± 1.35 |
2.494 ± 0.057 | 3.803 ± 0.301 |
2.431 ± 0.754 | 8.479 ± 1.339 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.12 ± 0.377 | 3.055 ± 0.113 |
6.297 ± 1.466 | 2.431 ± 0.184 |
7.481 ± 0.85 | 6.983 ± 0.436 |
6.359 ± 0.061 | 8.292 ± 0.022 |
1.434 ± 0.265 | 3.865 ± 0.934 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
