
Syntrophaceticus schinkii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Thermoanaerobacterales; Thermoanaerobacterales Family III. Incertae Sedis; Syntrophaceticus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
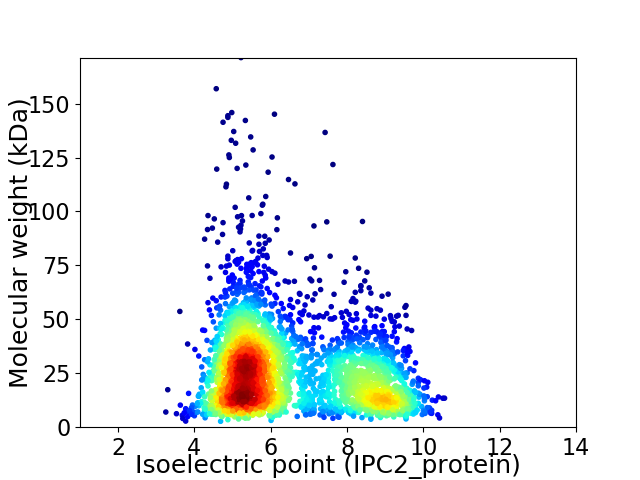
Virtual 2D-PAGE plot for 3167 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B7MQ13|A0A0B7MQ13_9FIRM Hydrogenase expression/formation protein HypC OS=Syntrophaceticus schinkii OX=499207 GN=hypC PE=3 SV=1
MM1 pKa = 7.88DD2 pKa = 5.9LVPEE6 pKa = 5.28HH7 pKa = 6.97ISSAYY12 pKa = 9.24AGGSIEE18 pKa = 4.0HH19 pKa = 5.99VKK21 pKa = 10.54VSRR24 pKa = 11.84EE25 pKa = 3.99SGEE28 pKa = 4.07SGNNSEE34 pKa = 5.76LILIEE39 pKa = 3.82SWGTYY44 pKa = 8.57QEE46 pKa = 4.38ACCILQAMIRR56 pKa = 11.84VDD58 pKa = 3.44RR59 pKa = 11.84SLDD62 pKa = 3.49FGRR65 pKa = 11.84GLCISSPSEE74 pKa = 3.97KK75 pKa = 10.08PSVFSPGCRR84 pKa = 11.84IISEE88 pKa = 4.67LYY90 pKa = 8.84NTDD93 pKa = 2.9GALNISDD100 pKa = 3.76STVNGDD106 pKa = 3.18IYY108 pKa = 10.99TGGDD112 pKa = 3.17LTMSGDD118 pKa = 3.18THH120 pKa = 7.21LAGDD124 pKa = 3.84INGEE128 pKa = 4.35GMFTACPNCRR138 pKa = 11.84VVGNVQVTGDD148 pKa = 3.71VQVEE152 pKa = 4.05DD153 pKa = 4.28DD154 pKa = 4.23VVIEE158 pKa = 3.91GDD160 pKa = 2.89IRR162 pKa = 11.84AAGDD166 pKa = 3.4VYY168 pKa = 10.66IRR170 pKa = 11.84KK171 pKa = 9.03AAVSGSIWSNGEE183 pKa = 3.51IFVEE187 pKa = 4.19QGGQVEE193 pKa = 4.4GGIYY197 pKa = 9.68PGQAMEE203 pKa = 5.42LDD205 pKa = 3.51VALPFFPEE213 pKa = 3.39VDD215 pKa = 3.08LSFFRR220 pKa = 11.84RR221 pKa = 11.84GADD224 pKa = 3.31QILKK228 pKa = 8.66GTQQLHH234 pKa = 6.8GILHH238 pKa = 6.58FDD240 pKa = 2.91GVTFIEE246 pKa = 4.94GDD248 pKa = 3.57LEE250 pKa = 4.1IAGDD254 pKa = 3.9YY255 pKa = 8.25TGKK258 pKa = 10.95GILVVDD264 pKa = 4.44GSVGISGDD272 pKa = 3.86LLPVGIQDD280 pKa = 3.99SLCILAAGPVTCADD294 pKa = 4.44GSCLSLLVYY303 pKa = 10.63GKK305 pKa = 10.45DD306 pKa = 3.48DD307 pKa = 3.91LSLGEE312 pKa = 3.94GSRR315 pKa = 11.84FQGSITAYY323 pKa = 8.08TLRR326 pKa = 11.84MCNNAEE332 pKa = 4.15FIYY335 pKa = 11.01DD336 pKa = 3.74GDD338 pKa = 4.36LVDD341 pKa = 4.75
MM1 pKa = 7.88DD2 pKa = 5.9LVPEE6 pKa = 5.28HH7 pKa = 6.97ISSAYY12 pKa = 9.24AGGSIEE18 pKa = 4.0HH19 pKa = 5.99VKK21 pKa = 10.54VSRR24 pKa = 11.84EE25 pKa = 3.99SGEE28 pKa = 4.07SGNNSEE34 pKa = 5.76LILIEE39 pKa = 3.82SWGTYY44 pKa = 8.57QEE46 pKa = 4.38ACCILQAMIRR56 pKa = 11.84VDD58 pKa = 3.44RR59 pKa = 11.84SLDD62 pKa = 3.49FGRR65 pKa = 11.84GLCISSPSEE74 pKa = 3.97KK75 pKa = 10.08PSVFSPGCRR84 pKa = 11.84IISEE88 pKa = 4.67LYY90 pKa = 8.84NTDD93 pKa = 2.9GALNISDD100 pKa = 3.76STVNGDD106 pKa = 3.18IYY108 pKa = 10.99TGGDD112 pKa = 3.17LTMSGDD118 pKa = 3.18THH120 pKa = 7.21LAGDD124 pKa = 3.84INGEE128 pKa = 4.35GMFTACPNCRR138 pKa = 11.84VVGNVQVTGDD148 pKa = 3.71VQVEE152 pKa = 4.05DD153 pKa = 4.28DD154 pKa = 4.23VVIEE158 pKa = 3.91GDD160 pKa = 2.89IRR162 pKa = 11.84AAGDD166 pKa = 3.4VYY168 pKa = 10.66IRR170 pKa = 11.84KK171 pKa = 9.03AAVSGSIWSNGEE183 pKa = 3.51IFVEE187 pKa = 4.19QGGQVEE193 pKa = 4.4GGIYY197 pKa = 9.68PGQAMEE203 pKa = 5.42LDD205 pKa = 3.51VALPFFPEE213 pKa = 3.39VDD215 pKa = 3.08LSFFRR220 pKa = 11.84RR221 pKa = 11.84GADD224 pKa = 3.31QILKK228 pKa = 8.66GTQQLHH234 pKa = 6.8GILHH238 pKa = 6.58FDD240 pKa = 2.91GVTFIEE246 pKa = 4.94GDD248 pKa = 3.57LEE250 pKa = 4.1IAGDD254 pKa = 3.9YY255 pKa = 8.25TGKK258 pKa = 10.95GILVVDD264 pKa = 4.44GSVGISGDD272 pKa = 3.86LLPVGIQDD280 pKa = 3.99SLCILAAGPVTCADD294 pKa = 4.44GSCLSLLVYY303 pKa = 10.63GKK305 pKa = 10.45DD306 pKa = 3.48DD307 pKa = 3.91LSLGEE312 pKa = 3.94GSRR315 pKa = 11.84FQGSITAYY323 pKa = 8.08TLRR326 pKa = 11.84MCNNAEE332 pKa = 4.15FIYY335 pKa = 11.01DD336 pKa = 3.74GDD338 pKa = 4.36LVDD341 pKa = 4.75
Molecular weight: 36.05 kDa
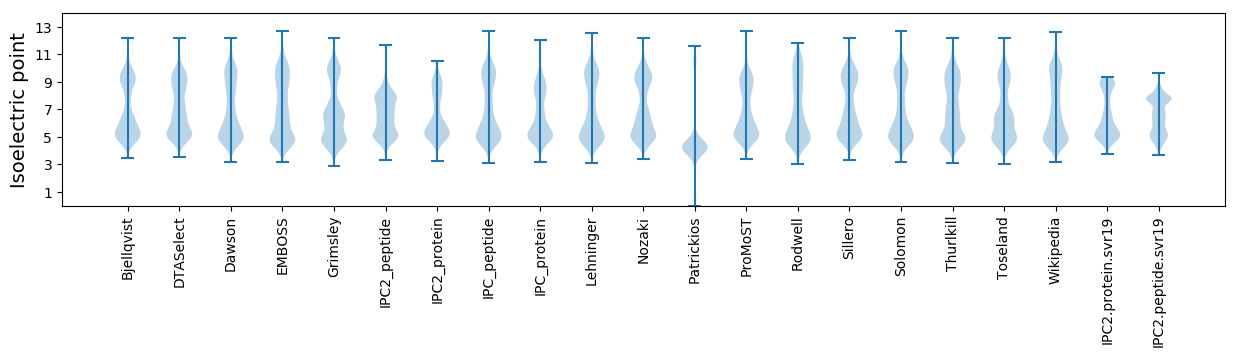
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B7MBM4|A0A0B7MBM4_9FIRM Uncharacterized protein OS=Syntrophaceticus schinkii OX=499207 GN=SSCH_1060007 PE=4 SV=1
MM1 pKa = 7.26TCLFAWNFGGIEE13 pKa = 3.91ITSLRR18 pKa = 11.84RR19 pKa = 11.84LDD21 pKa = 4.24VEE23 pKa = 4.29TQRR26 pKa = 11.84SQKK29 pKa = 10.53SLAEE33 pKa = 3.91IKK35 pKa = 9.65IWPARR40 pKa = 11.84EE41 pKa = 4.23FIYY44 pKa = 10.74HH45 pKa = 7.44PDD47 pKa = 3.17LAAPAVEE54 pKa = 5.39RR55 pKa = 11.84IQEE58 pKa = 4.16AYY60 pKa = 9.58KK61 pKa = 10.41KK62 pKa = 10.58RR63 pKa = 11.84RR64 pKa = 11.84DD65 pKa = 3.56ALKK68 pKa = 10.32GSRR71 pKa = 11.84NALTRR76 pKa = 11.84LQQRR80 pKa = 11.84ANRR83 pKa = 11.84FLEE86 pKa = 4.12MARR89 pKa = 11.84EE90 pKa = 4.31GVGRR94 pKa = 11.84SLNLVQPYY102 pKa = 8.75FYY104 pKa = 10.08PEE106 pKa = 3.92QVSLLDD112 pKa = 3.8YY113 pKa = 11.05LPADD117 pKa = 3.52SLVILDD123 pKa = 3.74EE124 pKa = 4.54PGRR127 pKa = 11.84FPEE130 pKa = 3.98QSRR133 pKa = 11.84ARR135 pKa = 11.84TVLLDD140 pKa = 3.17SDD142 pKa = 3.69YY143 pKa = 11.41RR144 pKa = 11.84RR145 pKa = 11.84FFQEE149 pKa = 4.36GNSFTPWQEE158 pKa = 3.82HH159 pKa = 5.79YY160 pKa = 11.04FDD162 pKa = 5.48GDD164 pKa = 3.49QLLKK168 pKa = 10.95QMAEE172 pKa = 4.19FPLICYY178 pKa = 9.93SRR180 pKa = 11.84LLTRR184 pKa = 11.84TPLMEE189 pKa = 4.87PKK191 pKa = 10.49ALVSFSTKK199 pKa = 7.62EE200 pKa = 3.55MRR202 pKa = 11.84PFLGRR207 pKa = 11.84PDD209 pKa = 4.96LLAQEE214 pKa = 4.38LKK216 pKa = 10.25GWLRR220 pKa = 11.84KK221 pKa = 8.45KK222 pKa = 7.47TTVLLLTHH230 pKa = 6.74TEE232 pKa = 4.22TNLKK236 pKa = 9.46QLEE239 pKa = 4.23RR240 pKa = 11.84EE241 pKa = 4.12LRR243 pKa = 11.84DD244 pKa = 3.38MEE246 pKa = 4.34ISFLIADD253 pKa = 3.77QWQEE257 pKa = 3.84CLKK260 pKa = 10.6EE261 pKa = 3.86GRR263 pKa = 11.84LQVGISSLSRR273 pKa = 11.84GFEE276 pKa = 3.71IPGILAVVSAMEE288 pKa = 3.99MYY290 pKa = 10.39GGKK293 pKa = 9.56KK294 pKa = 8.14VRR296 pKa = 11.84RR297 pKa = 11.84AQRR300 pKa = 11.84RR301 pKa = 11.84RR302 pKa = 11.84ATAADD307 pKa = 3.82HH308 pKa = 6.73LPEE311 pKa = 5.64LSLEE315 pKa = 4.43TLWCMSITGSGAMWASVRR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84QMISRR340 pKa = 11.84GITWRR345 pKa = 11.84SFMPGRR351 pKa = 11.84QGYY354 pKa = 7.79LCRR357 pKa = 11.84WIRR360 pKa = 11.84WTWFRR365 pKa = 11.84STQVRR370 pKa = 11.84KK371 pKa = 9.6GLHH374 pKa = 5.23RR375 pKa = 11.84VFPGWEE381 pKa = 3.9AVTGRR386 pKa = 11.84ASSSVSKK393 pKa = 10.67SAFRR397 pKa = 11.84SWLKK401 pKa = 10.66ACSLFMRR408 pKa = 11.84SAAMLQVMPFPRR420 pKa = 11.84ILSGSGSLRR429 pKa = 11.84MAFPMRR435 pKa = 11.84RR436 pKa = 11.84HH437 pKa = 7.19RR438 pKa = 11.84IRR440 pKa = 11.84KK441 pKa = 9.13RR442 pKa = 11.84PSEE445 pKa = 4.01MM446 pKa = 3.68
MM1 pKa = 7.26TCLFAWNFGGIEE13 pKa = 3.91ITSLRR18 pKa = 11.84RR19 pKa = 11.84LDD21 pKa = 4.24VEE23 pKa = 4.29TQRR26 pKa = 11.84SQKK29 pKa = 10.53SLAEE33 pKa = 3.91IKK35 pKa = 9.65IWPARR40 pKa = 11.84EE41 pKa = 4.23FIYY44 pKa = 10.74HH45 pKa = 7.44PDD47 pKa = 3.17LAAPAVEE54 pKa = 5.39RR55 pKa = 11.84IQEE58 pKa = 4.16AYY60 pKa = 9.58KK61 pKa = 10.41KK62 pKa = 10.58RR63 pKa = 11.84RR64 pKa = 11.84DD65 pKa = 3.56ALKK68 pKa = 10.32GSRR71 pKa = 11.84NALTRR76 pKa = 11.84LQQRR80 pKa = 11.84ANRR83 pKa = 11.84FLEE86 pKa = 4.12MARR89 pKa = 11.84EE90 pKa = 4.31GVGRR94 pKa = 11.84SLNLVQPYY102 pKa = 8.75FYY104 pKa = 10.08PEE106 pKa = 3.92QVSLLDD112 pKa = 3.8YY113 pKa = 11.05LPADD117 pKa = 3.52SLVILDD123 pKa = 3.74EE124 pKa = 4.54PGRR127 pKa = 11.84FPEE130 pKa = 3.98QSRR133 pKa = 11.84ARR135 pKa = 11.84TVLLDD140 pKa = 3.17SDD142 pKa = 3.69YY143 pKa = 11.41RR144 pKa = 11.84RR145 pKa = 11.84FFQEE149 pKa = 4.36GNSFTPWQEE158 pKa = 3.82HH159 pKa = 5.79YY160 pKa = 11.04FDD162 pKa = 5.48GDD164 pKa = 3.49QLLKK168 pKa = 10.95QMAEE172 pKa = 4.19FPLICYY178 pKa = 9.93SRR180 pKa = 11.84LLTRR184 pKa = 11.84TPLMEE189 pKa = 4.87PKK191 pKa = 10.49ALVSFSTKK199 pKa = 7.62EE200 pKa = 3.55MRR202 pKa = 11.84PFLGRR207 pKa = 11.84PDD209 pKa = 4.96LLAQEE214 pKa = 4.38LKK216 pKa = 10.25GWLRR220 pKa = 11.84KK221 pKa = 8.45KK222 pKa = 7.47TTVLLLTHH230 pKa = 6.74TEE232 pKa = 4.22TNLKK236 pKa = 9.46QLEE239 pKa = 4.23RR240 pKa = 11.84EE241 pKa = 4.12LRR243 pKa = 11.84DD244 pKa = 3.38MEE246 pKa = 4.34ISFLIADD253 pKa = 3.77QWQEE257 pKa = 3.84CLKK260 pKa = 10.6EE261 pKa = 3.86GRR263 pKa = 11.84LQVGISSLSRR273 pKa = 11.84GFEE276 pKa = 3.71IPGILAVVSAMEE288 pKa = 3.99MYY290 pKa = 10.39GGKK293 pKa = 9.56KK294 pKa = 8.14VRR296 pKa = 11.84RR297 pKa = 11.84AQRR300 pKa = 11.84RR301 pKa = 11.84RR302 pKa = 11.84ATAADD307 pKa = 3.82HH308 pKa = 6.73LPEE311 pKa = 5.64LSLEE315 pKa = 4.43TLWCMSITGSGAMWASVRR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84QMISRR340 pKa = 11.84GITWRR345 pKa = 11.84SFMPGRR351 pKa = 11.84QGYY354 pKa = 7.79LCRR357 pKa = 11.84WIRR360 pKa = 11.84WTWFRR365 pKa = 11.84STQVRR370 pKa = 11.84KK371 pKa = 9.6GLHH374 pKa = 5.23RR375 pKa = 11.84VFPGWEE381 pKa = 3.9AVTGRR386 pKa = 11.84ASSSVSKK393 pKa = 10.67SAFRR397 pKa = 11.84SWLKK401 pKa = 10.66ACSLFMRR408 pKa = 11.84SAAMLQVMPFPRR420 pKa = 11.84ILSGSGSLRR429 pKa = 11.84MAFPMRR435 pKa = 11.84RR436 pKa = 11.84HH437 pKa = 7.19RR438 pKa = 11.84IRR440 pKa = 11.84KK441 pKa = 9.13RR442 pKa = 11.84PSEE445 pKa = 4.01MM446 pKa = 3.68
Molecular weight: 51.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
794821 |
26 |
1487 |
251.0 |
27.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.849 ± 0.046 | 1.485 ± 0.023 |
5.224 ± 0.04 | 7.02 ± 0.051 |
4.003 ± 0.03 | 7.698 ± 0.052 |
1.85 ± 0.019 | 7.066 ± 0.045 |
5.897 ± 0.045 | 10.363 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.588 ± 0.022 | 3.611 ± 0.034 |
4.201 ± 0.031 | 3.517 ± 0.032 |
5.372 ± 0.042 | 5.544 ± 0.034 |
4.915 ± 0.036 | 7.411 ± 0.04 |
1.043 ± 0.018 | 3.343 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
