
Planctomycetes bacterium MalM25
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; unclassified Planctomycetes
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
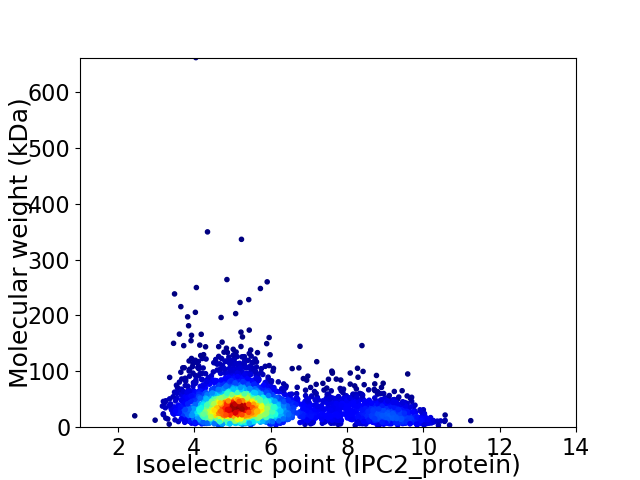
Virtual 2D-PAGE plot for 3738 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A517TGN6|A0A517TGN6_9BACT Uncharacterized protein OS=Planctomycetes bacterium MalM25 OX=2527998 GN=MalM25_04320 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 10.12SLCIALATCCSLALFQGTASALDD25 pKa = 3.49IYY27 pKa = 11.34VDD29 pKa = 3.91NFDD32 pKa = 4.96SYY34 pKa = 12.02DD35 pKa = 3.45AGSDD39 pKa = 2.78IGGPNWQPKK48 pKa = 7.43WAADD52 pKa = 3.56STQQALFKK60 pKa = 10.52AAAGGEE66 pKa = 4.65GYY68 pKa = 10.88GVIDD72 pKa = 3.28TTVAEE77 pKa = 3.83RR78 pKa = 11.84QYY80 pKa = 11.19RR81 pKa = 11.84VNGKK85 pKa = 9.87SGFSLLAGDD94 pKa = 4.46TVNVAADD101 pKa = 3.68FRR103 pKa = 11.84YY104 pKa = 10.0HH105 pKa = 7.72LEE107 pKa = 4.26AGGDD111 pKa = 3.53APAVFNSNVIGLQLSDD127 pKa = 4.95SEE129 pKa = 4.43NWWAGSRR136 pKa = 11.84KK137 pKa = 9.57SVSLANRR144 pKa = 11.84GPAMGNRR151 pKa = 11.84LPVAPWVQNWIPHH164 pKa = 5.62TSLGIADD171 pKa = 4.27PALGGISEE179 pKa = 4.86WININLEE186 pKa = 4.19LMVSDD191 pKa = 4.22GTALTTAGDD200 pKa = 4.34MIPAGNIYY208 pKa = 10.48GSATFTPEE216 pKa = 3.66SLAGEE221 pKa = 4.17PVEE224 pKa = 4.33NLPVTDD230 pKa = 4.64VLDD233 pKa = 4.18LGYY236 pKa = 8.52TAGTMLYY243 pKa = 10.71AGFTTDD249 pKa = 2.72WTSVAEE255 pKa = 4.31GEE257 pKa = 4.32PTTVSSYY264 pKa = 11.42ASVSEE269 pKa = 3.98VNIDD273 pKa = 3.6NFSLVSDD280 pKa = 4.47AEE282 pKa = 4.33APAGDD287 pKa = 4.21YY288 pKa = 10.93NQDD291 pKa = 3.43GYY293 pKa = 11.95VDD295 pKa = 3.63AADD298 pKa = 3.54YY299 pKa = 8.12TIWRR303 pKa = 11.84DD304 pKa = 3.29SNGQVGGGLAADD316 pKa = 4.02GTGDD320 pKa = 4.31DD321 pKa = 4.68LLGVPDD327 pKa = 4.66GDD329 pKa = 3.56VDD331 pKa = 4.67SFDD334 pKa = 4.17YY335 pKa = 11.2DD336 pKa = 3.2FWDD339 pKa = 3.53ANYY342 pKa = 10.58GSSAFSAPPVATAVPEE358 pKa = 4.55PGTLALVGFAALVAFRR374 pKa = 11.84RR375 pKa = 11.84RR376 pKa = 3.36
MM1 pKa = 7.66KK2 pKa = 10.12SLCIALATCCSLALFQGTASALDD25 pKa = 3.49IYY27 pKa = 11.34VDD29 pKa = 3.91NFDD32 pKa = 4.96SYY34 pKa = 12.02DD35 pKa = 3.45AGSDD39 pKa = 2.78IGGPNWQPKK48 pKa = 7.43WAADD52 pKa = 3.56STQQALFKK60 pKa = 10.52AAAGGEE66 pKa = 4.65GYY68 pKa = 10.88GVIDD72 pKa = 3.28TTVAEE77 pKa = 3.83RR78 pKa = 11.84QYY80 pKa = 11.19RR81 pKa = 11.84VNGKK85 pKa = 9.87SGFSLLAGDD94 pKa = 4.46TVNVAADD101 pKa = 3.68FRR103 pKa = 11.84YY104 pKa = 10.0HH105 pKa = 7.72LEE107 pKa = 4.26AGGDD111 pKa = 3.53APAVFNSNVIGLQLSDD127 pKa = 4.95SEE129 pKa = 4.43NWWAGSRR136 pKa = 11.84KK137 pKa = 9.57SVSLANRR144 pKa = 11.84GPAMGNRR151 pKa = 11.84LPVAPWVQNWIPHH164 pKa = 5.62TSLGIADD171 pKa = 4.27PALGGISEE179 pKa = 4.86WININLEE186 pKa = 4.19LMVSDD191 pKa = 4.22GTALTTAGDD200 pKa = 4.34MIPAGNIYY208 pKa = 10.48GSATFTPEE216 pKa = 3.66SLAGEE221 pKa = 4.17PVEE224 pKa = 4.33NLPVTDD230 pKa = 4.64VLDD233 pKa = 4.18LGYY236 pKa = 8.52TAGTMLYY243 pKa = 10.71AGFTTDD249 pKa = 2.72WTSVAEE255 pKa = 4.31GEE257 pKa = 4.32PTTVSSYY264 pKa = 11.42ASVSEE269 pKa = 3.98VNIDD273 pKa = 3.6NFSLVSDD280 pKa = 4.47AEE282 pKa = 4.33APAGDD287 pKa = 4.21YY288 pKa = 10.93NQDD291 pKa = 3.43GYY293 pKa = 11.95VDD295 pKa = 3.63AADD298 pKa = 3.54YY299 pKa = 8.12TIWRR303 pKa = 11.84DD304 pKa = 3.29SNGQVGGGLAADD316 pKa = 4.02GTGDD320 pKa = 4.31DD321 pKa = 4.68LLGVPDD327 pKa = 4.66GDD329 pKa = 3.56VDD331 pKa = 4.67SFDD334 pKa = 4.17YY335 pKa = 11.2DD336 pKa = 3.2FWDD339 pKa = 3.53ANYY342 pKa = 10.58GSSAFSAPPVATAVPEE358 pKa = 4.55PGTLALVGFAALVAFRR374 pKa = 11.84RR375 pKa = 11.84RR376 pKa = 3.36
Molecular weight: 39.27 kDa
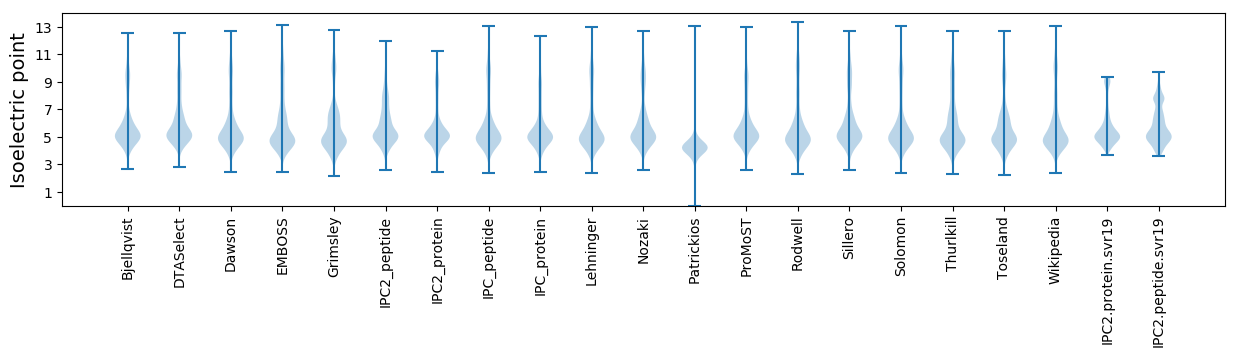
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A517TGM5|A0A517TGM5_9BACT Thioesterase (YiiD_Cterm) OS=Planctomycetes bacterium MalM25 OX=2527998 GN=MalM25_04290 PE=4 SV=1
MM1 pKa = 7.65LGFPLATALIVAGPAWASLGVALAAVAGFLAHH33 pKa = 7.01EE34 pKa = 4.49PLLVIRR40 pKa = 11.84GRR42 pKa = 11.84RR43 pKa = 11.84GLRR46 pKa = 11.84AQRR49 pKa = 11.84EE50 pKa = 4.22APAAKK55 pKa = 9.28QWLWAYY61 pKa = 10.79ALIATTGSSLAFGLGSSAVRR81 pKa = 11.84WSLLCCGAIATAGFALAWAGKK102 pKa = 7.58HH103 pKa = 5.73HH104 pKa = 6.61SRR106 pKa = 11.84AGQAFGAVGLSAPCVPILLSEE127 pKa = 4.37SEE129 pKa = 4.17ASIATAAEE137 pKa = 5.72FWATWLIGLAAAMIAVNSVLAPAKK161 pKa = 10.2RR162 pKa = 11.84LARR165 pKa = 11.84PTATGLLFLLGVATAILQAIGCHH188 pKa = 6.1LPMAVIPVFGLAMTLLLYY206 pKa = 9.75PPRR209 pKa = 11.84PKK211 pKa = 10.13HH212 pKa = 5.65LKK214 pKa = 9.35RR215 pKa = 11.84VGWTLAGGVFATSIWLLTLCC235 pKa = 5.25
MM1 pKa = 7.65LGFPLATALIVAGPAWASLGVALAAVAGFLAHH33 pKa = 7.01EE34 pKa = 4.49PLLVIRR40 pKa = 11.84GRR42 pKa = 11.84RR43 pKa = 11.84GLRR46 pKa = 11.84AQRR49 pKa = 11.84EE50 pKa = 4.22APAAKK55 pKa = 9.28QWLWAYY61 pKa = 10.79ALIATTGSSLAFGLGSSAVRR81 pKa = 11.84WSLLCCGAIATAGFALAWAGKK102 pKa = 7.58HH103 pKa = 5.73HH104 pKa = 6.61SRR106 pKa = 11.84AGQAFGAVGLSAPCVPILLSEE127 pKa = 4.37SEE129 pKa = 4.17ASIATAAEE137 pKa = 5.72FWATWLIGLAAAMIAVNSVLAPAKK161 pKa = 10.2RR162 pKa = 11.84LARR165 pKa = 11.84PTATGLLFLLGVATAILQAIGCHH188 pKa = 6.1LPMAVIPVFGLAMTLLLYY206 pKa = 9.75PPRR209 pKa = 11.84PKK211 pKa = 10.13HH212 pKa = 5.65LKK214 pKa = 9.35RR215 pKa = 11.84VGWTLAGGVFATSIWLLTLCC235 pKa = 5.25
Molecular weight: 24.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1380535 |
29 |
6256 |
369.3 |
40.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.367 ± 0.05 | 0.991 ± 0.017 |
6.361 ± 0.039 | 6.758 ± 0.044 |
3.355 ± 0.025 | 8.746 ± 0.041 |
1.997 ± 0.02 | 3.994 ± 0.024 |
2.947 ± 0.034 | 10.068 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.881 ± 0.016 | 2.755 ± 0.03 |
5.677 ± 0.03 | 3.307 ± 0.022 |
7.01 ± 0.046 | 5.798 ± 0.03 |
5.61 ± 0.032 | 7.375 ± 0.03 |
1.558 ± 0.018 | 2.444 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
