
Lake Sarah-associated circular virus-11
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.77
Get precalculated fractions of proteins
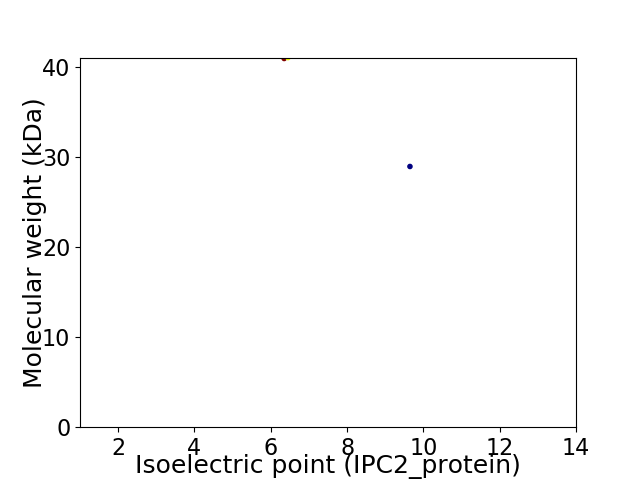
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
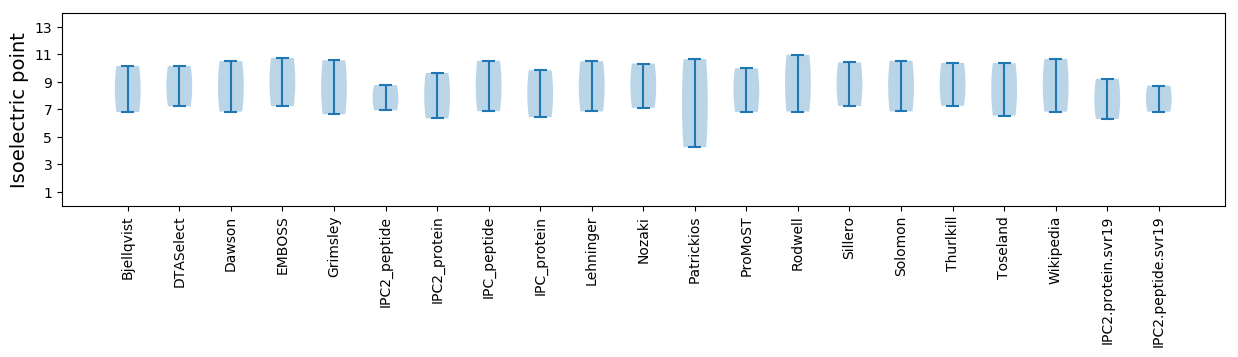
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126G9K5|A0A126G9K5_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-11 OX=1685737 PE=4 SV=1
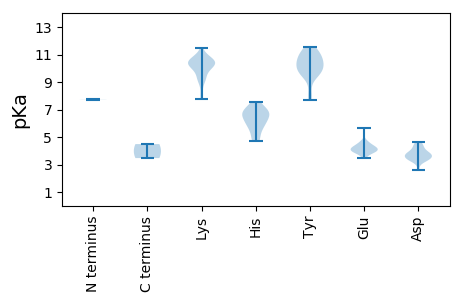
MM1 pKa = 7.74APRR4 pKa = 11.84GNAAKK9 pKa = 10.27RR10 pKa = 11.84WTFTINNWTLADD22 pKa = 3.76YY23 pKa = 11.19DD24 pKa = 3.97KK25 pKa = 11.47VLEE28 pKa = 4.29FTTKK32 pKa = 10.27FEE34 pKa = 4.48PVFLICGKK42 pKa = 10.02EE43 pKa = 3.84RR44 pKa = 11.84GEE46 pKa = 4.48LLTDD50 pKa = 3.68HH51 pKa = 6.7LQGFVHH57 pKa = 6.83LKK59 pKa = 9.61SKK61 pKa = 10.48QRR63 pKa = 11.84LTFLKK68 pKa = 10.79ANLSEE73 pKa = 4.18RR74 pKa = 11.84AHH76 pKa = 6.15FEE78 pKa = 3.93CAKK81 pKa = 10.84GSDD84 pKa = 3.82EE85 pKa = 5.66DD86 pKa = 4.09NRR88 pKa = 11.84DD89 pKa = 3.49YY90 pKa = 11.14CSKK93 pKa = 10.89EE94 pKa = 3.65DD95 pKa = 3.69TEE97 pKa = 4.37PFIFGTPVKK106 pKa = 10.36SCSEE110 pKa = 3.97KK111 pKa = 10.19GGGRR115 pKa = 11.84ITAQQVIGAAIALATEE131 pKa = 4.58RR132 pKa = 11.84EE133 pKa = 4.14PFLVIKK139 pKa = 10.45KK140 pKa = 9.66NEE142 pKa = 3.91EE143 pKa = 3.5FAVPVLRR150 pKa = 11.84HH151 pKa = 4.94YY152 pKa = 11.58ASFEE156 pKa = 3.8KK157 pKa = 10.61LAASVRR163 pKa = 11.84RR164 pKa = 11.84LNALEE169 pKa = 3.92QLQSEE174 pKa = 4.66YY175 pKa = 11.21KK176 pKa = 9.68EE177 pKa = 3.95VRR179 pKa = 11.84LKK181 pKa = 10.56VWQQAIINTIQEE193 pKa = 4.19RR194 pKa = 11.84PHH196 pKa = 6.64PRR198 pKa = 11.84TIHH201 pKa = 6.01WYY203 pKa = 8.2WEE205 pKa = 4.05ATGGTGKK212 pKa = 7.95TWMSKK217 pKa = 10.67YY218 pKa = 10.49LVACHH223 pKa = 5.76SAFRR227 pKa = 11.84AEE229 pKa = 4.16NGKK232 pKa = 10.83SNDD235 pKa = 2.63IKK237 pKa = 10.88YY238 pKa = 10.3AYY240 pKa = 10.01AGEE243 pKa = 4.2KK244 pKa = 10.34VVIFDD249 pKa = 3.58FTRR252 pKa = 11.84SQTEE256 pKa = 4.06HH257 pKa = 6.35INYY260 pKa = 9.83EE261 pKa = 3.91IIEE264 pKa = 4.42SIKK267 pKa = 10.26NGCYY271 pKa = 9.99FNCKK275 pKa = 9.24YY276 pKa = 10.23EE277 pKa = 4.1SGMRR281 pKa = 11.84IFPTPHH287 pKa = 5.68VFCFSNSPPDD297 pKa = 3.92LSKK300 pKa = 10.58MSADD304 pKa = 3.28RR305 pKa = 11.84WHH307 pKa = 6.89ISQIRR312 pKa = 11.84GLLLSWSVPLPIPGTRR328 pKa = 11.84TVDD331 pKa = 3.26EE332 pKa = 4.67SSVCEE337 pKa = 3.98TSSEE341 pKa = 4.2EE342 pKa = 4.13GGEE345 pKa = 3.91EE346 pKa = 4.04SFNLFEE352 pKa = 4.79YY353 pKa = 11.11LDD355 pKa = 3.66ATKK358 pKa = 9.96TT359 pKa = 3.47
MM1 pKa = 7.74APRR4 pKa = 11.84GNAAKK9 pKa = 10.27RR10 pKa = 11.84WTFTINNWTLADD22 pKa = 3.76YY23 pKa = 11.19DD24 pKa = 3.97KK25 pKa = 11.47VLEE28 pKa = 4.29FTTKK32 pKa = 10.27FEE34 pKa = 4.48PVFLICGKK42 pKa = 10.02EE43 pKa = 3.84RR44 pKa = 11.84GEE46 pKa = 4.48LLTDD50 pKa = 3.68HH51 pKa = 6.7LQGFVHH57 pKa = 6.83LKK59 pKa = 9.61SKK61 pKa = 10.48QRR63 pKa = 11.84LTFLKK68 pKa = 10.79ANLSEE73 pKa = 4.18RR74 pKa = 11.84AHH76 pKa = 6.15FEE78 pKa = 3.93CAKK81 pKa = 10.84GSDD84 pKa = 3.82EE85 pKa = 5.66DD86 pKa = 4.09NRR88 pKa = 11.84DD89 pKa = 3.49YY90 pKa = 11.14CSKK93 pKa = 10.89EE94 pKa = 3.65DD95 pKa = 3.69TEE97 pKa = 4.37PFIFGTPVKK106 pKa = 10.36SCSEE110 pKa = 3.97KK111 pKa = 10.19GGGRR115 pKa = 11.84ITAQQVIGAAIALATEE131 pKa = 4.58RR132 pKa = 11.84EE133 pKa = 4.14PFLVIKK139 pKa = 10.45KK140 pKa = 9.66NEE142 pKa = 3.91EE143 pKa = 3.5FAVPVLRR150 pKa = 11.84HH151 pKa = 4.94YY152 pKa = 11.58ASFEE156 pKa = 3.8KK157 pKa = 10.61LAASVRR163 pKa = 11.84RR164 pKa = 11.84LNALEE169 pKa = 3.92QLQSEE174 pKa = 4.66YY175 pKa = 11.21KK176 pKa = 9.68EE177 pKa = 3.95VRR179 pKa = 11.84LKK181 pKa = 10.56VWQQAIINTIQEE193 pKa = 4.19RR194 pKa = 11.84PHH196 pKa = 6.64PRR198 pKa = 11.84TIHH201 pKa = 6.01WYY203 pKa = 8.2WEE205 pKa = 4.05ATGGTGKK212 pKa = 7.95TWMSKK217 pKa = 10.67YY218 pKa = 10.49LVACHH223 pKa = 5.76SAFRR227 pKa = 11.84AEE229 pKa = 4.16NGKK232 pKa = 10.83SNDD235 pKa = 2.63IKK237 pKa = 10.88YY238 pKa = 10.3AYY240 pKa = 10.01AGEE243 pKa = 4.2KK244 pKa = 10.34VVIFDD249 pKa = 3.58FTRR252 pKa = 11.84SQTEE256 pKa = 4.06HH257 pKa = 6.35INYY260 pKa = 9.83EE261 pKa = 3.91IIEE264 pKa = 4.42SIKK267 pKa = 10.26NGCYY271 pKa = 9.99FNCKK275 pKa = 9.24YY276 pKa = 10.23EE277 pKa = 4.1SGMRR281 pKa = 11.84IFPTPHH287 pKa = 5.68VFCFSNSPPDD297 pKa = 3.92LSKK300 pKa = 10.58MSADD304 pKa = 3.28RR305 pKa = 11.84WHH307 pKa = 6.89ISQIRR312 pKa = 11.84GLLLSWSVPLPIPGTRR328 pKa = 11.84TVDD331 pKa = 3.26EE332 pKa = 4.67SSVCEE337 pKa = 3.98TSSEE341 pKa = 4.2EE342 pKa = 4.13GGEE345 pKa = 3.91EE346 pKa = 4.04SFNLFEE352 pKa = 4.79YY353 pKa = 11.11LDD355 pKa = 3.66ATKK358 pKa = 9.96TT359 pKa = 3.47
Molecular weight: 40.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126G9K5|A0A126G9K5_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-11 OX=1685737 PE=4 SV=1
MM1 pKa = 7.73KK2 pKa = 10.09YY3 pKa = 10.64ASRR6 pKa = 11.84KK7 pKa = 9.19GRR9 pKa = 11.84GKK11 pKa = 10.37RR12 pKa = 11.84KK13 pKa = 8.37STTSHH18 pKa = 6.9RR19 pKa = 11.84GRR21 pKa = 11.84GGKK24 pKa = 7.78VRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84LFRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 9.07SFLKK38 pKa = 10.38RR39 pKa = 11.84RR40 pKa = 11.84TGNGGFANNTARR52 pKa = 11.84RR53 pKa = 11.84YY54 pKa = 10.37KK55 pKa = 10.51KK56 pKa = 10.58GLTAKK61 pKa = 10.2IVGDD65 pKa = 4.24DD66 pKa = 3.45FTLAIEE72 pKa = 4.35AAGGDD77 pKa = 4.52SNTMLIMYY85 pKa = 9.61DD86 pKa = 3.47GGNLGVSQFAMQQINAYY103 pKa = 7.67STLYY107 pKa = 10.7DD108 pKa = 3.46SYY110 pKa = 10.97KK111 pKa = 9.31VKK113 pKa = 10.55RR114 pKa = 11.84VVAEE118 pKa = 3.6WWLNDD123 pKa = 3.21NDD125 pKa = 4.27EE126 pKa = 4.23LAKK129 pKa = 10.43PDD131 pKa = 3.75TQLVNMWQCYY141 pKa = 9.5DD142 pKa = 3.4HH143 pKa = 7.53DD144 pKa = 4.61AFSKK148 pKa = 10.2VLKK151 pKa = 9.94IDD153 pKa = 4.41SIKK156 pKa = 9.96QQPGVKK162 pKa = 9.1RR163 pKa = 11.84HH164 pKa = 4.73IMMPGRR170 pKa = 11.84VYY172 pKa = 9.69KK173 pKa = 10.54TSFVPKK179 pKa = 10.05FVVNNKK185 pKa = 8.98NNQYY189 pKa = 9.63MEE191 pKa = 4.44GFNTWMDD198 pKa = 3.74LKK200 pKa = 11.18DD201 pKa = 3.34IAKK204 pKa = 9.5TNASGNALQFGWEE217 pKa = 4.27GKK219 pKa = 10.37AEE221 pKa = 3.86EE222 pKa = 4.27AAISVRR228 pKa = 11.84ITYY231 pKa = 9.41YY232 pKa = 10.16FAFGGRR238 pKa = 11.84RR239 pKa = 11.84VNSVYY244 pKa = 10.66QNSVDD249 pKa = 3.85NTVHH253 pKa = 6.88LL254 pKa = 4.49
MM1 pKa = 7.73KK2 pKa = 10.09YY3 pKa = 10.64ASRR6 pKa = 11.84KK7 pKa = 9.19GRR9 pKa = 11.84GKK11 pKa = 10.37RR12 pKa = 11.84KK13 pKa = 8.37STTSHH18 pKa = 6.9RR19 pKa = 11.84GRR21 pKa = 11.84GGKK24 pKa = 7.78VRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84LFRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 9.07SFLKK38 pKa = 10.38RR39 pKa = 11.84RR40 pKa = 11.84TGNGGFANNTARR52 pKa = 11.84RR53 pKa = 11.84YY54 pKa = 10.37KK55 pKa = 10.51KK56 pKa = 10.58GLTAKK61 pKa = 10.2IVGDD65 pKa = 4.24DD66 pKa = 3.45FTLAIEE72 pKa = 4.35AAGGDD77 pKa = 4.52SNTMLIMYY85 pKa = 9.61DD86 pKa = 3.47GGNLGVSQFAMQQINAYY103 pKa = 7.67STLYY107 pKa = 10.7DD108 pKa = 3.46SYY110 pKa = 10.97KK111 pKa = 9.31VKK113 pKa = 10.55RR114 pKa = 11.84VVAEE118 pKa = 3.6WWLNDD123 pKa = 3.21NDD125 pKa = 4.27EE126 pKa = 4.23LAKK129 pKa = 10.43PDD131 pKa = 3.75TQLVNMWQCYY141 pKa = 9.5DD142 pKa = 3.4HH143 pKa = 7.53DD144 pKa = 4.61AFSKK148 pKa = 10.2VLKK151 pKa = 9.94IDD153 pKa = 4.41SIKK156 pKa = 9.96QQPGVKK162 pKa = 9.1RR163 pKa = 11.84HH164 pKa = 4.73IMMPGRR170 pKa = 11.84VYY172 pKa = 9.69KK173 pKa = 10.54TSFVPKK179 pKa = 10.05FVVNNKK185 pKa = 8.98NNQYY189 pKa = 9.63MEE191 pKa = 4.44GFNTWMDD198 pKa = 3.74LKK200 pKa = 11.18DD201 pKa = 3.34IAKK204 pKa = 9.5TNASGNALQFGWEE217 pKa = 4.27GKK219 pKa = 10.37AEE221 pKa = 3.86EE222 pKa = 4.27AAISVRR228 pKa = 11.84ITYY231 pKa = 9.41YY232 pKa = 10.16FAFGGRR238 pKa = 11.84RR239 pKa = 11.84VNSVYY244 pKa = 10.66QNSVDD249 pKa = 3.85NTVHH253 pKa = 6.88LL254 pKa = 4.49
Molecular weight: 28.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
613 |
254 |
359 |
306.5 |
34.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.341 ± 0.09 | 1.631 ± 0.798 |
4.405 ± 0.714 | 6.525 ± 2.429 |
5.383 ± 0.425 | 7.178 ± 1.21 |
2.284 ± 0.457 | 5.057 ± 0.722 |
7.83 ± 0.789 | 6.525 ± 0.653 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.121 ± 0.917 | 5.546 ± 1.246 |
3.1 ± 0.983 | 3.426 ± 0.329 |
6.688 ± 1.018 | 6.852 ± 0.61 |
6.199 ± 0.443 | 5.873 ± 0.782 |
2.121 ± 0.098 | 3.915 ± 0.522 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
