
Trailing lespedeza virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae
Average proteome isoelectric point is 7.99
Get precalculated fractions of proteins
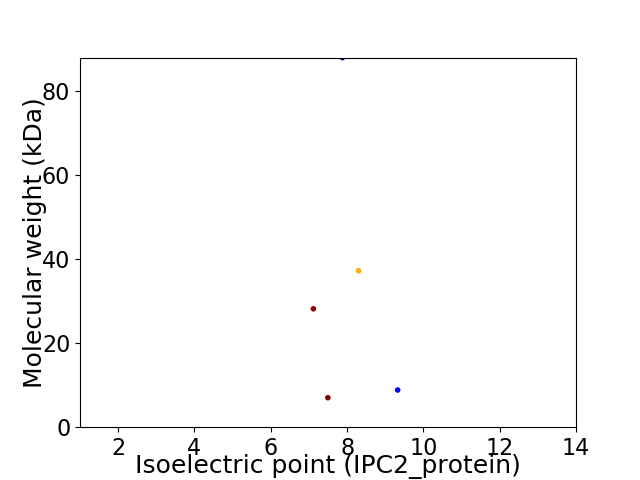
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F1BA31|F1BA31_9TOMB Putative movement protein 1 OS=Trailing lespedeza virus 1 OX=944580 PE=4 SV=1
MM1 pKa = 7.4FRR3 pKa = 11.84DD4 pKa = 3.95LCRR7 pKa = 11.84YY8 pKa = 9.27ASQIVVGSTVLAVNVSVLSVAVQLRR33 pKa = 11.84GAVAVYY39 pKa = 9.09EE40 pKa = 4.35VASKK44 pKa = 10.84CVTALEE50 pKa = 4.72DD51 pKa = 3.86CVSDD55 pKa = 5.13LRR57 pKa = 11.84DD58 pKa = 3.47SLKK61 pKa = 10.51PVPDD65 pKa = 3.51EE66 pKa = 4.71LYY68 pKa = 11.09NPSLCAALRR77 pKa = 11.84MNNTDD82 pKa = 3.52TEE84 pKa = 4.41LTAEE88 pKa = 4.71DD89 pKa = 5.31LLTTDD94 pKa = 3.62RR95 pKa = 11.84VLVNTWEE102 pKa = 3.92TDD104 pKa = 3.21EE105 pKa = 5.84AKK107 pKa = 10.43FKK109 pKa = 10.78YY110 pKa = 9.63RR111 pKa = 11.84YY112 pKa = 8.79KK113 pKa = 11.02ARR115 pKa = 11.84IQSGQRR121 pKa = 11.84GRR123 pKa = 11.84FMYY126 pKa = 10.48EE127 pKa = 3.51LVAYY131 pKa = 8.46GKK133 pKa = 9.46NHH135 pKa = 6.76FGRR138 pKa = 11.84APRR141 pKa = 11.84CCDD144 pKa = 3.2SNYY147 pKa = 10.52LSVTRR152 pKa = 11.84LLVNRR157 pKa = 11.84CNEE160 pKa = 3.81LHH162 pKa = 6.16LTPTQTRR169 pKa = 11.84NVVAEE174 pKa = 3.99AVSLVFIPDD183 pKa = 3.41EE184 pKa = 4.15EE185 pKa = 4.78EE186 pKa = 3.64IRR188 pKa = 11.84QTMLTNSHH196 pKa = 5.7YY197 pKa = 11.04AYY199 pKa = 9.23MQRR202 pKa = 11.84TRR204 pKa = 11.84QSVVSTVEE212 pKa = 3.96PWWKK216 pKa = 10.2EE217 pKa = 3.7LLLGPSNAKK226 pKa = 8.27NWRR229 pKa = 11.84RR230 pKa = 11.84AWIEE234 pKa = 3.63ICGLPEE240 pKa = 3.64KK241 pKa = 10.39QAISFAKK248 pKa = 10.36
MM1 pKa = 7.4FRR3 pKa = 11.84DD4 pKa = 3.95LCRR7 pKa = 11.84YY8 pKa = 9.27ASQIVVGSTVLAVNVSVLSVAVQLRR33 pKa = 11.84GAVAVYY39 pKa = 9.09EE40 pKa = 4.35VASKK44 pKa = 10.84CVTALEE50 pKa = 4.72DD51 pKa = 3.86CVSDD55 pKa = 5.13LRR57 pKa = 11.84DD58 pKa = 3.47SLKK61 pKa = 10.51PVPDD65 pKa = 3.51EE66 pKa = 4.71LYY68 pKa = 11.09NPSLCAALRR77 pKa = 11.84MNNTDD82 pKa = 3.52TEE84 pKa = 4.41LTAEE88 pKa = 4.71DD89 pKa = 5.31LLTTDD94 pKa = 3.62RR95 pKa = 11.84VLVNTWEE102 pKa = 3.92TDD104 pKa = 3.21EE105 pKa = 5.84AKK107 pKa = 10.43FKK109 pKa = 10.78YY110 pKa = 9.63RR111 pKa = 11.84YY112 pKa = 8.79KK113 pKa = 11.02ARR115 pKa = 11.84IQSGQRR121 pKa = 11.84GRR123 pKa = 11.84FMYY126 pKa = 10.48EE127 pKa = 3.51LVAYY131 pKa = 8.46GKK133 pKa = 9.46NHH135 pKa = 6.76FGRR138 pKa = 11.84APRR141 pKa = 11.84CCDD144 pKa = 3.2SNYY147 pKa = 10.52LSVTRR152 pKa = 11.84LLVNRR157 pKa = 11.84CNEE160 pKa = 3.81LHH162 pKa = 6.16LTPTQTRR169 pKa = 11.84NVVAEE174 pKa = 3.99AVSLVFIPDD183 pKa = 3.41EE184 pKa = 4.15EE185 pKa = 4.78EE186 pKa = 3.64IRR188 pKa = 11.84QTMLTNSHH196 pKa = 5.7YY197 pKa = 11.04AYY199 pKa = 9.23MQRR202 pKa = 11.84TRR204 pKa = 11.84QSVVSTVEE212 pKa = 3.96PWWKK216 pKa = 10.2EE217 pKa = 3.7LLLGPSNAKK226 pKa = 8.27NWRR229 pKa = 11.84RR230 pKa = 11.84AWIEE234 pKa = 3.63ICGLPEE240 pKa = 3.64KK241 pKa = 10.39QAISFAKK248 pKa = 10.36
Molecular weight: 28.19 kDa
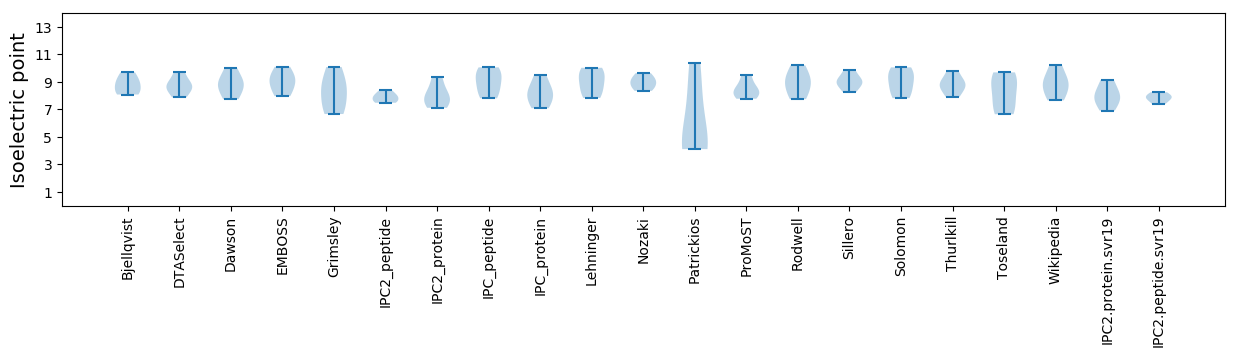
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F1BA33|F1BA33_9TOMB Capsid protein OS=Trailing lespedeza virus 1 OX=944580 PE=3 SV=1
MM1 pKa = 6.87SHH3 pKa = 6.38SRR5 pKa = 11.84GLIHH9 pKa = 6.82TLLVLLLVVSLLWLKK24 pKa = 9.69QYY26 pKa = 10.22RR27 pKa = 11.84WSITLTFDD35 pKa = 2.98SSLNRR40 pKa = 11.84VIALALLGAVLTSVGNSSTVYY61 pKa = 10.55YY62 pKa = 10.28INNSTNEE69 pKa = 3.8NKK71 pKa = 9.62TNHH74 pKa = 5.86IKK76 pKa = 10.34IEE78 pKa = 4.17TT79 pKa = 3.68
MM1 pKa = 6.87SHH3 pKa = 6.38SRR5 pKa = 11.84GLIHH9 pKa = 6.82TLLVLLLVVSLLWLKK24 pKa = 9.69QYY26 pKa = 10.22RR27 pKa = 11.84WSITLTFDD35 pKa = 2.98SSLNRR40 pKa = 11.84VIALALLGAVLTSVGNSSTVYY61 pKa = 10.55YY62 pKa = 10.28INNSTNEE69 pKa = 3.8NKK71 pKa = 9.62TNHH74 pKa = 5.86IKK76 pKa = 10.34IEE78 pKa = 4.17TT79 pKa = 3.68
Molecular weight: 8.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1510 |
67 |
770 |
302.0 |
33.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.079 ± 0.766 | 2.583 ± 0.548 |
4.437 ± 0.341 | 5.099 ± 0.899 |
3.444 ± 0.492 | 5.497 ± 0.721 |
2.185 ± 0.76 | 4.636 ± 0.657 |
4.437 ± 0.206 | 8.874 ± 1.376 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.921 ± 0.508 | 5.364 ± 0.387 |
3.841 ± 0.471 | 3.311 ± 0.295 |
6.954 ± 0.717 | 7.483 ± 1.42 |
7.02 ± 1.216 | 9.073 ± 0.822 |
1.854 ± 0.141 | 3.841 ± 0.472 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
