
Lysinimonas sp. KACC 19322
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Protaetiibacter
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
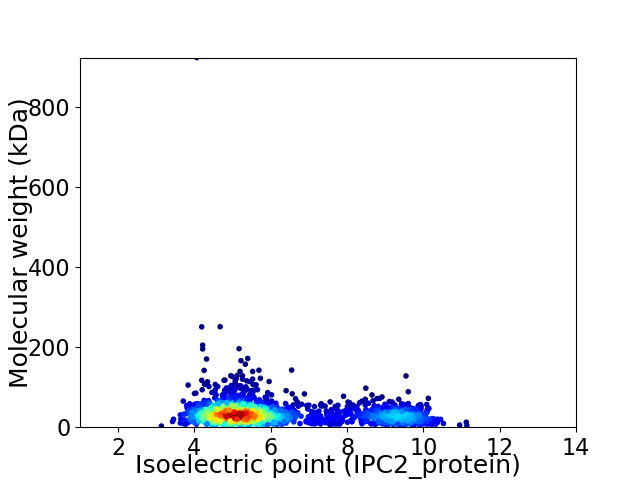
Virtual 2D-PAGE plot for 2341 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C1Y5L6|A0A5C1Y5L6_9MICO Uncharacterized protein OS=Lysinimonas sp. KACC 19322 OX=2592654 GN=FLP23_03645 PE=4 SV=1
MM1 pKa = 7.28SASPTSRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84SYY12 pKa = 11.5AGIAVAAFLATALTACFPGGGDD34 pKa = 3.42SKK36 pKa = 11.52SGGGVSYY43 pKa = 10.92DD44 pKa = 4.0GEE46 pKa = 4.69DD47 pKa = 3.37SAAVQSATIQFSGQGTFVEE66 pKa = 5.19PDD68 pKa = 3.4TIDD71 pKa = 3.49PAEE74 pKa = 4.04YY75 pKa = 8.97RR76 pKa = 11.84WSGSGFFITSDD87 pKa = 3.99GIAVTNNHH95 pKa = 5.51VVTGAGSLSVTIGGEE110 pKa = 3.98GDD112 pKa = 3.72SYY114 pKa = 10.64PVRR117 pKa = 11.84VLSTSEE123 pKa = 4.02CLDD126 pKa = 3.33IAVVQVDD133 pKa = 3.17VDD135 pKa = 3.79KK136 pKa = 10.81PVPYY140 pKa = 10.35FDD142 pKa = 3.34WAEE145 pKa = 4.11GKK147 pKa = 10.13ISTGSDD153 pKa = 3.49VYY155 pKa = 11.69ALGYY159 pKa = 9.59PLGDD163 pKa = 3.57PTFSMTKK170 pKa = 9.21GAVVKK175 pKa = 10.44SDD177 pKa = 3.4YY178 pKa = 11.68AFDD181 pKa = 3.65TFDD184 pKa = 3.32NVVAHH189 pKa = 7.2GIQSDD194 pKa = 3.52AAIRR198 pKa = 11.84GGNSGGPLINEE209 pKa = 4.06KK210 pKa = 10.8GKK212 pKa = 10.03VVGVNFYY219 pKa = 11.58GNDD222 pKa = 3.23NLATSNYY229 pKa = 10.33AIGRR233 pKa = 11.84DD234 pKa = 3.59DD235 pKa = 4.19VLPVLDD241 pKa = 4.84DD242 pKa = 4.34LKK244 pKa = 11.05AGKK247 pKa = 9.82SVLSLGLNLSALTTDD262 pKa = 4.09DD263 pKa = 4.47PYY265 pKa = 11.47GLSGIWVQSVAEE277 pKa = 4.56GSAADD282 pKa = 3.77KK283 pKa = 11.24AGIEE287 pKa = 4.37PGDD290 pKa = 3.76VLQKK294 pKa = 10.99LGGVTLAGASSLSQFEE310 pKa = 4.35NTTEE314 pKa = 3.99GTMKK318 pKa = 10.54NYY320 pKa = 10.39CQVLQTKK327 pKa = 10.24GVDD330 pKa = 3.35APMGAVVYY338 pKa = 10.49RR339 pKa = 11.84PATDD343 pKa = 3.27EE344 pKa = 4.11LLEE347 pKa = 4.16GEE349 pKa = 4.86INGSDD354 pKa = 4.22PIEE357 pKa = 4.15VTASGVLGGGGDD369 pKa = 3.8GGNVSSGYY377 pKa = 9.14TDD379 pKa = 3.08IVSDD383 pKa = 4.27DD384 pKa = 3.32YY385 pKa = 11.37RR386 pKa = 11.84IAVTVPAEE394 pKa = 4.06WNQVATTPGAGEE406 pKa = 4.21TLLQASSDD414 pKa = 3.62LGSFGSLAAPGVEE427 pKa = 4.18LHH429 pKa = 5.95STDD432 pKa = 3.8YY433 pKa = 10.8QALGAPADD441 pKa = 4.05VIATLDD447 pKa = 3.85LGLTNLGCTPSSGQIDD463 pKa = 3.61QDD465 pKa = 4.13YY466 pKa = 11.69ADD468 pKa = 4.1GQYY471 pKa = 10.97SGLFSVYY478 pKa = 9.36TSCPTGADD486 pKa = 2.95AVVVVVDD493 pKa = 4.87ADD495 pKa = 3.51NGDD498 pKa = 3.24ASGYY502 pKa = 10.77LLILGTFGDD511 pKa = 4.58YY512 pKa = 10.82ASSDD516 pKa = 3.54PGVLKK521 pKa = 10.81VLNTFTLSS529 pKa = 3.14
MM1 pKa = 7.28SASPTSRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84SYY12 pKa = 11.5AGIAVAAFLATALTACFPGGGDD34 pKa = 3.42SKK36 pKa = 11.52SGGGVSYY43 pKa = 10.92DD44 pKa = 4.0GEE46 pKa = 4.69DD47 pKa = 3.37SAAVQSATIQFSGQGTFVEE66 pKa = 5.19PDD68 pKa = 3.4TIDD71 pKa = 3.49PAEE74 pKa = 4.04YY75 pKa = 8.97RR76 pKa = 11.84WSGSGFFITSDD87 pKa = 3.99GIAVTNNHH95 pKa = 5.51VVTGAGSLSVTIGGEE110 pKa = 3.98GDD112 pKa = 3.72SYY114 pKa = 10.64PVRR117 pKa = 11.84VLSTSEE123 pKa = 4.02CLDD126 pKa = 3.33IAVVQVDD133 pKa = 3.17VDD135 pKa = 3.79KK136 pKa = 10.81PVPYY140 pKa = 10.35FDD142 pKa = 3.34WAEE145 pKa = 4.11GKK147 pKa = 10.13ISTGSDD153 pKa = 3.49VYY155 pKa = 11.69ALGYY159 pKa = 9.59PLGDD163 pKa = 3.57PTFSMTKK170 pKa = 9.21GAVVKK175 pKa = 10.44SDD177 pKa = 3.4YY178 pKa = 11.68AFDD181 pKa = 3.65TFDD184 pKa = 3.32NVVAHH189 pKa = 7.2GIQSDD194 pKa = 3.52AAIRR198 pKa = 11.84GGNSGGPLINEE209 pKa = 4.06KK210 pKa = 10.8GKK212 pKa = 10.03VVGVNFYY219 pKa = 11.58GNDD222 pKa = 3.23NLATSNYY229 pKa = 10.33AIGRR233 pKa = 11.84DD234 pKa = 3.59DD235 pKa = 4.19VLPVLDD241 pKa = 4.84DD242 pKa = 4.34LKK244 pKa = 11.05AGKK247 pKa = 9.82SVLSLGLNLSALTTDD262 pKa = 4.09DD263 pKa = 4.47PYY265 pKa = 11.47GLSGIWVQSVAEE277 pKa = 4.56GSAADD282 pKa = 3.77KK283 pKa = 11.24AGIEE287 pKa = 4.37PGDD290 pKa = 3.76VLQKK294 pKa = 10.99LGGVTLAGASSLSQFEE310 pKa = 4.35NTTEE314 pKa = 3.99GTMKK318 pKa = 10.54NYY320 pKa = 10.39CQVLQTKK327 pKa = 10.24GVDD330 pKa = 3.35APMGAVVYY338 pKa = 10.49RR339 pKa = 11.84PATDD343 pKa = 3.27EE344 pKa = 4.11LLEE347 pKa = 4.16GEE349 pKa = 4.86INGSDD354 pKa = 4.22PIEE357 pKa = 4.15VTASGVLGGGGDD369 pKa = 3.8GGNVSSGYY377 pKa = 9.14TDD379 pKa = 3.08IVSDD383 pKa = 4.27DD384 pKa = 3.32YY385 pKa = 11.37RR386 pKa = 11.84IAVTVPAEE394 pKa = 4.06WNQVATTPGAGEE406 pKa = 4.21TLLQASSDD414 pKa = 3.62LGSFGSLAAPGVEE427 pKa = 4.18LHH429 pKa = 5.95STDD432 pKa = 3.8YY433 pKa = 10.8QALGAPADD441 pKa = 4.05VIATLDD447 pKa = 3.85LGLTNLGCTPSSGQIDD463 pKa = 3.61QDD465 pKa = 4.13YY466 pKa = 11.69ADD468 pKa = 4.1GQYY471 pKa = 10.97SGLFSVYY478 pKa = 9.36TSCPTGADD486 pKa = 2.95AVVVVVDD493 pKa = 4.87ADD495 pKa = 3.51NGDD498 pKa = 3.24ASGYY502 pKa = 10.77LLILGTFGDD511 pKa = 4.58YY512 pKa = 10.82ASSDD516 pKa = 3.54PGVLKK521 pKa = 10.81VLNTFTLSS529 pKa = 3.14
Molecular weight: 53.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C1YCP6|A0A5C1YCP6_9MICO ABC transporter ATP-binding protein OS=Lysinimonas sp. KACC 19322 OX=2592654 GN=FLP23_10960 PE=4 SV=1
MM1 pKa = 7.44TIKK4 pKa = 10.42PSRR7 pKa = 11.84VSVRR11 pKa = 11.84PRR13 pKa = 11.84PTGRR17 pKa = 11.84PIRR20 pKa = 11.84GHH22 pKa = 5.36RR23 pKa = 11.84PRR25 pKa = 11.84RR26 pKa = 11.84PRR28 pKa = 11.84GPGRR32 pKa = 11.84RR33 pKa = 11.84GRR35 pKa = 11.84ARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84GGRR43 pKa = 11.84GTRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84GRR50 pKa = 11.84GQRR53 pKa = 11.84WGSWVVQTKK62 pKa = 10.09RR63 pKa = 11.84EE64 pKa = 4.09PGRR67 pKa = 11.84DD68 pKa = 3.02AGATSRR74 pKa = 11.84EE75 pKa = 4.21ATGLGLWARR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84GVAAAWRR93 pKa = 11.84GRR95 pKa = 11.84PGRR98 pKa = 11.84TRR100 pKa = 11.84AGLRR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84HH107 pKa = 5.92RR108 pKa = 11.84SS109 pKa = 2.98
MM1 pKa = 7.44TIKK4 pKa = 10.42PSRR7 pKa = 11.84VSVRR11 pKa = 11.84PRR13 pKa = 11.84PTGRR17 pKa = 11.84PIRR20 pKa = 11.84GHH22 pKa = 5.36RR23 pKa = 11.84PRR25 pKa = 11.84RR26 pKa = 11.84PRR28 pKa = 11.84GPGRR32 pKa = 11.84RR33 pKa = 11.84GRR35 pKa = 11.84ARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84GGRR43 pKa = 11.84GTRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84GRR50 pKa = 11.84GQRR53 pKa = 11.84WGSWVVQTKK62 pKa = 10.09RR63 pKa = 11.84EE64 pKa = 4.09PGRR67 pKa = 11.84DD68 pKa = 3.02AGATSRR74 pKa = 11.84EE75 pKa = 4.21ATGLGLWARR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84GVAAAWRR93 pKa = 11.84GRR95 pKa = 11.84PGRR98 pKa = 11.84TRR100 pKa = 11.84AGLRR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84HH107 pKa = 5.92RR108 pKa = 11.84SS109 pKa = 2.98
Molecular weight: 12.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
777487 |
26 |
9623 |
332.1 |
35.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.014 ± 0.1 | 0.437 ± 0.012 |
5.917 ± 0.042 | 5.79 ± 0.068 |
3.13 ± 0.035 | 9.16 ± 0.076 |
1.895 ± 0.026 | 4.521 ± 0.038 |
1.874 ± 0.037 | 10.471 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.438 ± 0.023 | 1.841 ± 0.025 |
5.591 ± 0.053 | 2.638 ± 0.03 |
7.539 ± 0.086 | 5.355 ± 0.055 |
5.806 ± 0.082 | 9.101 ± 0.053 |
1.504 ± 0.023 | 1.975 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
