
Altererythrobacter soli
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae; Croceibacterium
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
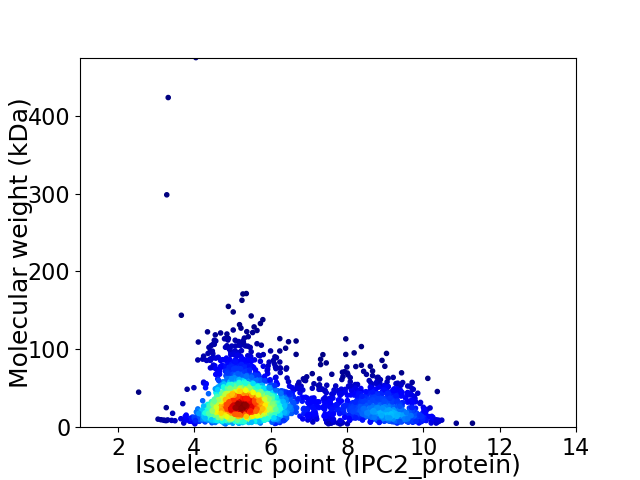
Virtual 2D-PAGE plot for 2807 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I4UX55|A0A6I4UX55_9SPHN DUF1178 family protein OS=Altererythrobacter soli OX=1739690 GN=GRI75_11885 PE=4 SV=1
MM1 pKa = 7.3SLSAKK6 pKa = 10.07SRR8 pKa = 11.84LLATSAVAGLCVWQTHH24 pKa = 5.71LPAAAQDD31 pKa = 3.45AFGIHH36 pKa = 6.59VDD38 pKa = 3.62TPEE41 pKa = 4.02GQVIVIEE48 pKa = 4.16NGEE51 pKa = 4.08IVEE54 pKa = 4.08GDD56 pKa = 3.64RR57 pKa = 11.84IGVYY61 pKa = 10.35AEE63 pKa = 4.0QGALDD68 pKa = 3.86LTVEE72 pKa = 4.17TGARR76 pKa = 11.84VRR78 pKa = 11.84GNGFYY83 pKa = 10.61DD84 pKa = 3.94GFGAPPEE91 pKa = 4.33AGVTIATAGSSVDD104 pKa = 3.4NSGSISGAGAGITTAYY120 pKa = 10.35VYY122 pKa = 11.05DD123 pKa = 3.98SAADD127 pKa = 3.67MLVGQAIGTVVVNNGTIAGDD147 pKa = 3.67SNDD150 pKa = 3.22GVRR153 pKa = 11.84LIGGGSVTNSGTILGANWDD172 pKa = 3.97FADD175 pKa = 5.4GISMYY180 pKa = 10.76AYY182 pKa = 10.2ADD184 pKa = 3.57QAKK187 pKa = 9.3EE188 pKa = 4.1DD189 pKa = 3.92YY190 pKa = 10.33SALVTNEE197 pKa = 3.71AGGEE201 pKa = 3.87IAGQRR206 pKa = 11.84FGVILSGGGDD216 pKa = 3.57VVNDD220 pKa = 3.63GDD222 pKa = 3.83ITGVAGGVLVQGTALDD238 pKa = 3.74SEE240 pKa = 4.57EE241 pKa = 4.85RR242 pKa = 11.84SGLTASVVNSGTISGTGSFGGTYY265 pKa = 9.87TGGQGVAFGSDD276 pKa = 2.83MAAATLVNSGTITSAFAEE294 pKa = 4.36AVSQGSLADD303 pKa = 3.54LTITNEE309 pKa = 3.66QSGVIEE315 pKa = 4.9GGTSGIYY322 pKa = 10.19GGSSGTMSIINAGTIRR338 pKa = 11.84GNGSYY343 pKa = 10.5DD344 pKa = 3.46GFGAPPDD351 pKa = 3.76AGITIATAGSSVDD364 pKa = 3.36NSGTISGAGAGITTAYY380 pKa = 10.33VYY382 pKa = 11.04DD383 pKa = 4.04SATDD387 pKa = 3.6TLVGQAIGTVVVNSGTIAGDD407 pKa = 3.51SNDD410 pKa = 3.22GVRR413 pKa = 11.84LIGGGSVTNSGTILGANWDD432 pKa = 3.97FADD435 pKa = 5.4GISMYY440 pKa = 10.76AYY442 pKa = 10.2ADD444 pKa = 3.57QAKK447 pKa = 9.3EE448 pKa = 4.1DD449 pKa = 3.92YY450 pKa = 10.33SALVTNEE457 pKa = 3.71AGGEE461 pKa = 3.87IAGQRR466 pKa = 11.84FGVILSGGGDD476 pKa = 3.57VVNDD480 pKa = 3.63GDD482 pKa = 3.83ITGVAGGVLVQGTALDD498 pKa = 3.74SEE500 pKa = 4.57EE501 pKa = 4.85RR502 pKa = 11.84SGLTASVVNSGTISGTGSLASGVAFGSDD530 pKa = 2.86MAAATLVNSGTINGAAAGIEE550 pKa = 4.51AYY552 pKa = 10.53FSGAFSLANTGTVTGEE568 pKa = 4.23SIGVDD573 pKa = 3.31SGALGAVTLNNSGSITGEE591 pKa = 3.69SGPAVVSASQTTLINSGSLHH611 pKa = 5.96GAGGVAVQLAEE622 pKa = 4.18YY623 pKa = 9.74DD624 pKa = 3.64DD625 pKa = 5.01SVTLRR630 pKa = 11.84TGSSVVGTIDD640 pKa = 4.36AGDD643 pKa = 3.8GTDD646 pKa = 3.5ILILEE651 pKa = 4.54GDD653 pKa = 4.08VLGLTSTQVIGSAEE667 pKa = 3.91NFEE670 pKa = 4.5TLRR673 pKa = 11.84VATGYY678 pKa = 7.1WTTASYY684 pKa = 11.24VGDD687 pKa = 4.18FEE689 pKa = 4.72QVAILEE695 pKa = 4.57GASLQVNEE703 pKa = 4.47VTVDD707 pKa = 3.52AQVTSPILTSDD718 pKa = 3.69VLNDD722 pKa = 3.49GTLVLNFDD730 pKa = 3.59HH731 pKa = 8.52VEE733 pKa = 3.79IDD735 pKa = 3.38EE736 pKa = 4.35LAISGSGGVTLIGEE750 pKa = 4.51AVYY753 pKa = 10.31EE754 pKa = 4.19VTGNSLTYY762 pKa = 10.48TGDD765 pKa = 3.33TVIANGGVILSGSLDD780 pKa = 3.65SDD782 pKa = 3.94VVTQGDD788 pKa = 4.12GFFTLGLGGTAGTYY802 pKa = 9.9VGSIVNNGVFNFDD815 pKa = 3.33RR816 pKa = 11.84SDD818 pKa = 3.3NYY820 pKa = 10.93EE821 pKa = 3.83FLGDD825 pKa = 3.79FSGSGTLNKK834 pKa = 10.09FGAGTLTFSGLYY846 pKa = 10.43DD847 pKa = 3.87FGGTTTINAGAIAFAGQLAEE867 pKa = 4.43DD868 pKa = 4.1TQLDD872 pKa = 3.97LSEE875 pKa = 5.45GGTFDD880 pKa = 4.94LSQIEE885 pKa = 4.51SGEE888 pKa = 3.83QTIAEE893 pKa = 4.08LSGTGGTLLLGDD905 pKa = 4.05TDD907 pKa = 3.96LTVQQTADD915 pKa = 3.53SVFAGAVTGTGTLLKK930 pKa = 10.74DD931 pKa = 3.52GSGDD935 pKa = 3.54LKK937 pKa = 11.42LNGDD941 pKa = 3.71GTGFTGTGQVDD952 pKa = 3.8GGTLSVNGDD961 pKa = 3.52FSNANFLVNDD971 pKa = 4.32GGTLGGAGTLGSTTVSGGTLAPGNSIDD998 pKa = 3.88TITFVGDD1005 pKa = 3.56LTLTAASVYY1014 pKa = 9.51EE1015 pKa = 4.28VEE1017 pKa = 4.67VDD1019 pKa = 3.36PAGNSDD1025 pKa = 3.55RR1026 pKa = 11.84TNVTGTATLGNATVNVLAEE1045 pKa = 4.18SGLYY1049 pKa = 10.48GPATDD1054 pKa = 3.97YY1055 pKa = 11.14TILTAEE1061 pKa = 4.51NGISGQFGEE1070 pKa = 4.59VQTNLAYY1077 pKa = 10.44LVPLLSYY1084 pKa = 10.23TVDD1087 pKa = 3.41TVSLRR1092 pKa = 11.84LSRR1095 pKa = 11.84NDD1097 pKa = 3.13IDD1099 pKa = 3.93FSTYY1103 pKa = 10.96AATPNQATVGALIEE1117 pKa = 4.24GLEE1120 pKa = 4.41FGTPLYY1126 pKa = 10.92DD1127 pKa = 3.26ATLVLARR1134 pKa = 11.84DD1135 pKa = 3.56QVATNFEE1142 pKa = 4.46TLTGGVYY1149 pKa = 7.91PTYY1152 pKa = 10.41GASLVEE1158 pKa = 3.83TAYY1161 pKa = 10.16MLRR1164 pKa = 11.84RR1165 pKa = 11.84QAAPQPQAAVGAFVWGTGLAGEE1187 pKa = 4.99LSASGSDD1194 pKa = 3.1GVTGFKK1200 pKa = 10.0TDD1202 pKa = 3.07NAGAAGGLGYY1212 pKa = 10.67AADD1215 pKa = 4.17GFDD1218 pKa = 3.56FTVGLGKK1225 pKa = 10.48LWQDD1229 pKa = 3.37PDD1231 pKa = 3.4RR1232 pKa = 11.84SQISDD1237 pKa = 3.57SDD1239 pKa = 3.92VKK1241 pKa = 10.79FALAQASYY1249 pKa = 10.45ASTMGLSARR1258 pKa = 11.84AGVQFGWIDD1267 pKa = 3.41GAASRR1272 pKa = 11.84NTRR1275 pKa = 11.84LGTISATVSSAFEE1288 pKa = 4.04GEE1290 pKa = 4.03YY1291 pKa = 9.42TSYY1294 pKa = 10.94YY1295 pKa = 10.77GEE1297 pKa = 4.28IGYY1300 pKa = 9.78ALPMGVASIEE1310 pKa = 3.99PFAGLNHH1317 pKa = 6.12VSLDD1321 pKa = 3.21MDD1323 pKa = 3.98RR1324 pKa = 11.84FTEE1327 pKa = 4.23VGGATALTVDD1337 pKa = 3.65GLDD1340 pKa = 3.51RR1341 pKa = 11.84DD1342 pKa = 3.97VTFADD1347 pKa = 3.52LGLRR1351 pKa = 11.84LAANPVGGVAPYY1363 pKa = 10.72LSGAYY1368 pKa = 8.89RR1369 pKa = 11.84RR1370 pKa = 11.84AWGDD1374 pKa = 3.16RR1375 pKa = 11.84MSEE1378 pKa = 3.72ATIAFTGNSAGGTITGLPIARR1399 pKa = 11.84SAAEE1403 pKa = 3.89LEE1405 pKa = 3.98AGVRR1409 pKa = 11.84YY1410 pKa = 10.27SGGVVDD1416 pKa = 6.24LEE1418 pKa = 4.3LGYY1421 pKa = 9.35TGSFSKK1427 pKa = 9.73TFDD1430 pKa = 3.61SNSVKK1435 pKa = 10.36VSASIHH1441 pKa = 5.03FF1442 pKa = 4.14
MM1 pKa = 7.3SLSAKK6 pKa = 10.07SRR8 pKa = 11.84LLATSAVAGLCVWQTHH24 pKa = 5.71LPAAAQDD31 pKa = 3.45AFGIHH36 pKa = 6.59VDD38 pKa = 3.62TPEE41 pKa = 4.02GQVIVIEE48 pKa = 4.16NGEE51 pKa = 4.08IVEE54 pKa = 4.08GDD56 pKa = 3.64RR57 pKa = 11.84IGVYY61 pKa = 10.35AEE63 pKa = 4.0QGALDD68 pKa = 3.86LTVEE72 pKa = 4.17TGARR76 pKa = 11.84VRR78 pKa = 11.84GNGFYY83 pKa = 10.61DD84 pKa = 3.94GFGAPPEE91 pKa = 4.33AGVTIATAGSSVDD104 pKa = 3.4NSGSISGAGAGITTAYY120 pKa = 10.35VYY122 pKa = 11.05DD123 pKa = 3.98SAADD127 pKa = 3.67MLVGQAIGTVVVNNGTIAGDD147 pKa = 3.67SNDD150 pKa = 3.22GVRR153 pKa = 11.84LIGGGSVTNSGTILGANWDD172 pKa = 3.97FADD175 pKa = 5.4GISMYY180 pKa = 10.76AYY182 pKa = 10.2ADD184 pKa = 3.57QAKK187 pKa = 9.3EE188 pKa = 4.1DD189 pKa = 3.92YY190 pKa = 10.33SALVTNEE197 pKa = 3.71AGGEE201 pKa = 3.87IAGQRR206 pKa = 11.84FGVILSGGGDD216 pKa = 3.57VVNDD220 pKa = 3.63GDD222 pKa = 3.83ITGVAGGVLVQGTALDD238 pKa = 3.74SEE240 pKa = 4.57EE241 pKa = 4.85RR242 pKa = 11.84SGLTASVVNSGTISGTGSFGGTYY265 pKa = 9.87TGGQGVAFGSDD276 pKa = 2.83MAAATLVNSGTITSAFAEE294 pKa = 4.36AVSQGSLADD303 pKa = 3.54LTITNEE309 pKa = 3.66QSGVIEE315 pKa = 4.9GGTSGIYY322 pKa = 10.19GGSSGTMSIINAGTIRR338 pKa = 11.84GNGSYY343 pKa = 10.5DD344 pKa = 3.46GFGAPPDD351 pKa = 3.76AGITIATAGSSVDD364 pKa = 3.36NSGTISGAGAGITTAYY380 pKa = 10.33VYY382 pKa = 11.04DD383 pKa = 4.04SATDD387 pKa = 3.6TLVGQAIGTVVVNSGTIAGDD407 pKa = 3.51SNDD410 pKa = 3.22GVRR413 pKa = 11.84LIGGGSVTNSGTILGANWDD432 pKa = 3.97FADD435 pKa = 5.4GISMYY440 pKa = 10.76AYY442 pKa = 10.2ADD444 pKa = 3.57QAKK447 pKa = 9.3EE448 pKa = 4.1DD449 pKa = 3.92YY450 pKa = 10.33SALVTNEE457 pKa = 3.71AGGEE461 pKa = 3.87IAGQRR466 pKa = 11.84FGVILSGGGDD476 pKa = 3.57VVNDD480 pKa = 3.63GDD482 pKa = 3.83ITGVAGGVLVQGTALDD498 pKa = 3.74SEE500 pKa = 4.57EE501 pKa = 4.85RR502 pKa = 11.84SGLTASVVNSGTISGTGSLASGVAFGSDD530 pKa = 2.86MAAATLVNSGTINGAAAGIEE550 pKa = 4.51AYY552 pKa = 10.53FSGAFSLANTGTVTGEE568 pKa = 4.23SIGVDD573 pKa = 3.31SGALGAVTLNNSGSITGEE591 pKa = 3.69SGPAVVSASQTTLINSGSLHH611 pKa = 5.96GAGGVAVQLAEE622 pKa = 4.18YY623 pKa = 9.74DD624 pKa = 3.64DD625 pKa = 5.01SVTLRR630 pKa = 11.84TGSSVVGTIDD640 pKa = 4.36AGDD643 pKa = 3.8GTDD646 pKa = 3.5ILILEE651 pKa = 4.54GDD653 pKa = 4.08VLGLTSTQVIGSAEE667 pKa = 3.91NFEE670 pKa = 4.5TLRR673 pKa = 11.84VATGYY678 pKa = 7.1WTTASYY684 pKa = 11.24VGDD687 pKa = 4.18FEE689 pKa = 4.72QVAILEE695 pKa = 4.57GASLQVNEE703 pKa = 4.47VTVDD707 pKa = 3.52AQVTSPILTSDD718 pKa = 3.69VLNDD722 pKa = 3.49GTLVLNFDD730 pKa = 3.59HH731 pKa = 8.52VEE733 pKa = 3.79IDD735 pKa = 3.38EE736 pKa = 4.35LAISGSGGVTLIGEE750 pKa = 4.51AVYY753 pKa = 10.31EE754 pKa = 4.19VTGNSLTYY762 pKa = 10.48TGDD765 pKa = 3.33TVIANGGVILSGSLDD780 pKa = 3.65SDD782 pKa = 3.94VVTQGDD788 pKa = 4.12GFFTLGLGGTAGTYY802 pKa = 9.9VGSIVNNGVFNFDD815 pKa = 3.33RR816 pKa = 11.84SDD818 pKa = 3.3NYY820 pKa = 10.93EE821 pKa = 3.83FLGDD825 pKa = 3.79FSGSGTLNKK834 pKa = 10.09FGAGTLTFSGLYY846 pKa = 10.43DD847 pKa = 3.87FGGTTTINAGAIAFAGQLAEE867 pKa = 4.43DD868 pKa = 4.1TQLDD872 pKa = 3.97LSEE875 pKa = 5.45GGTFDD880 pKa = 4.94LSQIEE885 pKa = 4.51SGEE888 pKa = 3.83QTIAEE893 pKa = 4.08LSGTGGTLLLGDD905 pKa = 4.05TDD907 pKa = 3.96LTVQQTADD915 pKa = 3.53SVFAGAVTGTGTLLKK930 pKa = 10.74DD931 pKa = 3.52GSGDD935 pKa = 3.54LKK937 pKa = 11.42LNGDD941 pKa = 3.71GTGFTGTGQVDD952 pKa = 3.8GGTLSVNGDD961 pKa = 3.52FSNANFLVNDD971 pKa = 4.32GGTLGGAGTLGSTTVSGGTLAPGNSIDD998 pKa = 3.88TITFVGDD1005 pKa = 3.56LTLTAASVYY1014 pKa = 9.51EE1015 pKa = 4.28VEE1017 pKa = 4.67VDD1019 pKa = 3.36PAGNSDD1025 pKa = 3.55RR1026 pKa = 11.84TNVTGTATLGNATVNVLAEE1045 pKa = 4.18SGLYY1049 pKa = 10.48GPATDD1054 pKa = 3.97YY1055 pKa = 11.14TILTAEE1061 pKa = 4.51NGISGQFGEE1070 pKa = 4.59VQTNLAYY1077 pKa = 10.44LVPLLSYY1084 pKa = 10.23TVDD1087 pKa = 3.41TVSLRR1092 pKa = 11.84LSRR1095 pKa = 11.84NDD1097 pKa = 3.13IDD1099 pKa = 3.93FSTYY1103 pKa = 10.96AATPNQATVGALIEE1117 pKa = 4.24GLEE1120 pKa = 4.41FGTPLYY1126 pKa = 10.92DD1127 pKa = 3.26ATLVLARR1134 pKa = 11.84DD1135 pKa = 3.56QVATNFEE1142 pKa = 4.46TLTGGVYY1149 pKa = 7.91PTYY1152 pKa = 10.41GASLVEE1158 pKa = 3.83TAYY1161 pKa = 10.16MLRR1164 pKa = 11.84RR1165 pKa = 11.84QAAPQPQAAVGAFVWGTGLAGEE1187 pKa = 4.99LSASGSDD1194 pKa = 3.1GVTGFKK1200 pKa = 10.0TDD1202 pKa = 3.07NAGAAGGLGYY1212 pKa = 10.67AADD1215 pKa = 4.17GFDD1218 pKa = 3.56FTVGLGKK1225 pKa = 10.48LWQDD1229 pKa = 3.37PDD1231 pKa = 3.4RR1232 pKa = 11.84SQISDD1237 pKa = 3.57SDD1239 pKa = 3.92VKK1241 pKa = 10.79FALAQASYY1249 pKa = 10.45ASTMGLSARR1258 pKa = 11.84AGVQFGWIDD1267 pKa = 3.41GAASRR1272 pKa = 11.84NTRR1275 pKa = 11.84LGTISATVSSAFEE1288 pKa = 4.04GEE1290 pKa = 4.03YY1291 pKa = 9.42TSYY1294 pKa = 10.94YY1295 pKa = 10.77GEE1297 pKa = 4.28IGYY1300 pKa = 9.78ALPMGVASIEE1310 pKa = 3.99PFAGLNHH1317 pKa = 6.12VSLDD1321 pKa = 3.21MDD1323 pKa = 3.98RR1324 pKa = 11.84FTEE1327 pKa = 4.23VGGATALTVDD1337 pKa = 3.65GLDD1340 pKa = 3.51RR1341 pKa = 11.84DD1342 pKa = 3.97VTFADD1347 pKa = 3.52LGLRR1351 pKa = 11.84LAANPVGGVAPYY1363 pKa = 10.72LSGAYY1368 pKa = 8.89RR1369 pKa = 11.84RR1370 pKa = 11.84AWGDD1374 pKa = 3.16RR1375 pKa = 11.84MSEE1378 pKa = 3.72ATIAFTGNSAGGTITGLPIARR1399 pKa = 11.84SAAEE1403 pKa = 3.89LEE1405 pKa = 3.98AGVRR1409 pKa = 11.84YY1410 pKa = 10.27SGGVVDD1416 pKa = 6.24LEE1418 pKa = 4.3LGYY1421 pKa = 9.35TGSFSKK1427 pKa = 9.73TFDD1430 pKa = 3.61SNSVKK1435 pKa = 10.36VSASIHH1441 pKa = 5.03FF1442 pKa = 4.14
Molecular weight: 143.75 kDa
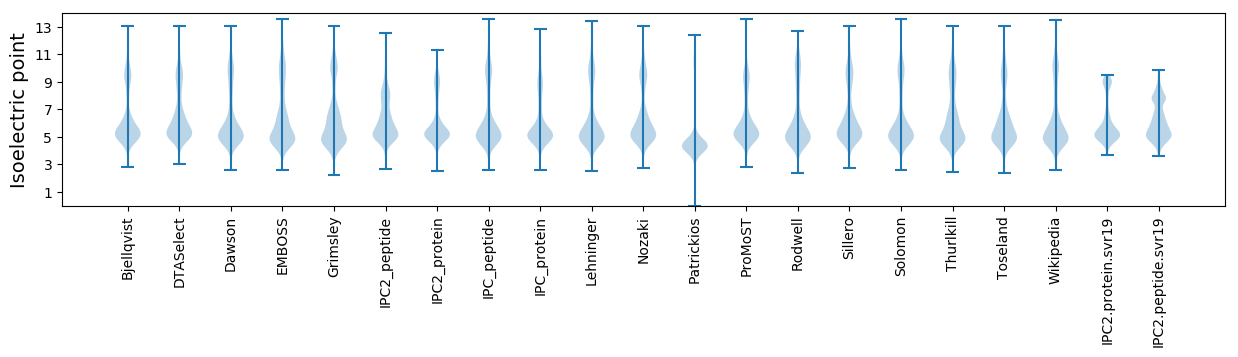
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I4UPF9|A0A6I4UPF9_9SPHN Uncharacterized protein OS=Altererythrobacter soli OX=1739690 GN=GRI75_04005 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84SRR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.5GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 8.12VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84SRR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.5GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 8.12VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
910999 |
38 |
4886 |
324.5 |
35.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.047 ± 0.071 | 0.816 ± 0.017 |
5.795 ± 0.039 | 6.414 ± 0.062 |
3.564 ± 0.029 | 9.118 ± 0.081 |
1.932 ± 0.026 | 4.662 ± 0.025 |
2.798 ± 0.046 | 9.938 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.305 ± 0.026 | 2.471 ± 0.039 |
5.443 ± 0.041 | 3.221 ± 0.025 |
7.438 ± 0.066 | 5.21 ± 0.043 |
5.092 ± 0.076 | 7.147 ± 0.038 |
1.412 ± 0.02 | 2.177 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
