
Paenibacillus sp. TCA20
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Paenibacillus; unclassified Paenibacillus
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
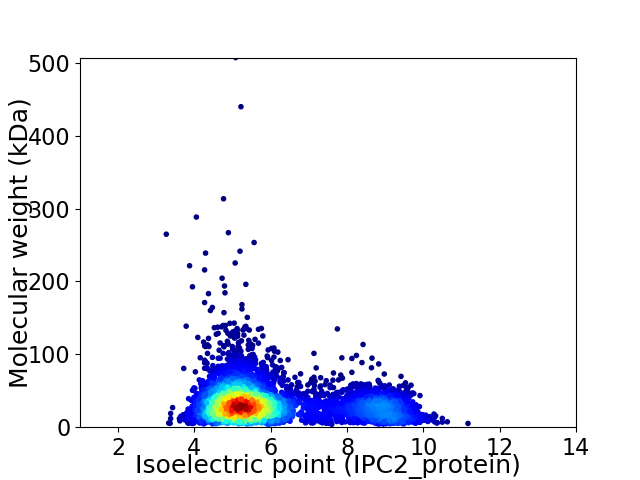
Virtual 2D-PAGE plot for 5226 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
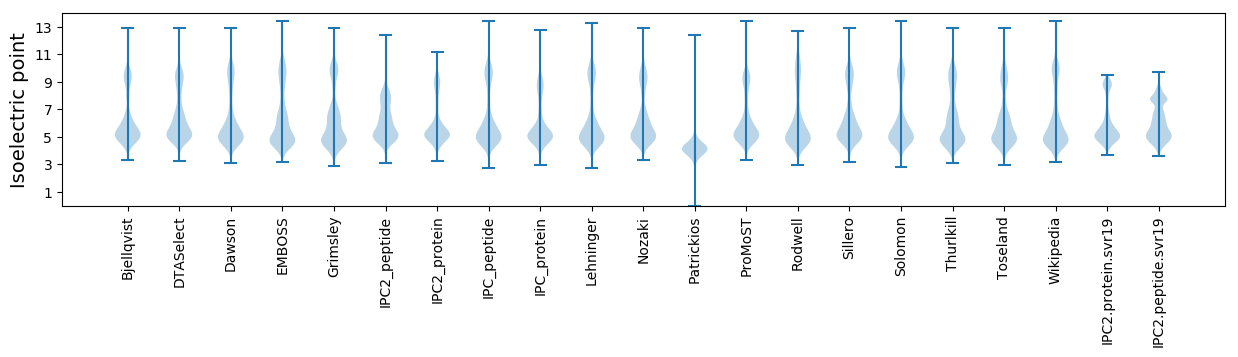
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A069DAF2|A0A069DAF2_9BACL UPF0060 membrane protein TCA2_1843 OS=Paenibacillus sp. TCA20 OX=1499968 GN=TCA2_1843 PE=3 SV=1
MM1 pKa = 6.85QSKK4 pKa = 9.78PIRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84LMSLSVVSTLFLSTVFPAVVSGAEE33 pKa = 3.7ATNRR37 pKa = 11.84KK38 pKa = 8.94FDD40 pKa = 3.94PSLFNLDD47 pKa = 3.38EE48 pKa = 4.76SKK50 pKa = 11.16LNSQMNGVTLNSSKK64 pKa = 10.66KK65 pKa = 9.4QAALNNTDD73 pKa = 2.7QSTNTGSKK81 pKa = 9.64IEE83 pKa = 5.09SYY85 pKa = 8.95TQSDD89 pKa = 3.34NSEE92 pKa = 4.1RR93 pKa = 11.84ASSVITPQNYY103 pKa = 8.99VPEE106 pKa = 4.65SDD108 pKa = 3.61STSTISVIVEE118 pKa = 3.95LSEE121 pKa = 4.2EE122 pKa = 4.53PIALYY127 pKa = 10.15EE128 pKa = 4.47SKK130 pKa = 10.71SSQNLNSGNNDD141 pKa = 2.99QRR143 pKa = 11.84SLIQKK148 pKa = 7.16QQQSFASAAKK158 pKa = 10.2RR159 pKa = 11.84LNATLKK165 pKa = 9.01TPYY168 pKa = 10.15SVVFNGYY175 pKa = 10.13AVDD178 pKa = 3.45IRR180 pKa = 11.84ADD182 pKa = 3.8RR183 pKa = 11.84VEE185 pKa = 4.36SLLAIPGVKK194 pKa = 9.52AVYY197 pKa = 9.59PDD199 pKa = 4.26SEE201 pKa = 4.48MKK203 pKa = 9.38ATPINSITPNMDD215 pKa = 2.56EE216 pKa = 4.12SAPFIGSDD224 pKa = 3.54TFWDD228 pKa = 3.09IGYY231 pKa = 9.67DD232 pKa = 3.45GAGIKK237 pKa = 10.5VGVLDD242 pKa = 3.48TGIDD246 pKa = 3.75YY247 pKa = 7.96EE248 pKa = 4.58HH249 pKa = 7.6PSLADD254 pKa = 3.69AYY256 pKa = 10.02KK257 pKa = 10.64GGYY260 pKa = 10.07DD261 pKa = 5.42FIDD264 pKa = 4.08NDD266 pKa = 4.21DD267 pKa = 4.94DD268 pKa = 4.75PYY270 pKa = 10.84EE271 pKa = 4.44TPPDD275 pKa = 4.27PNDD278 pKa = 3.55PEE280 pKa = 4.44AATDD284 pKa = 3.42HH285 pKa = 5.97GTHH288 pKa = 5.88VSGTIAGRR296 pKa = 11.84GNPLDD301 pKa = 4.36PEE303 pKa = 4.69SGTGWVRR310 pKa = 11.84GVAYY314 pKa = 10.04GADD317 pKa = 2.85IYY319 pKa = 10.95AYY321 pKa = 9.87RR322 pKa = 11.84VLGPGGSGPTSGVIAAIEE340 pKa = 3.89QSVEE344 pKa = 3.81DD345 pKa = 5.38DD346 pKa = 3.17MDD348 pKa = 5.01IINLSLGSEE357 pKa = 4.4MNNEE361 pKa = 4.06FDD363 pKa = 4.01PASVALNNAALAGVVAVVANGNSGPDD389 pKa = 3.92DD390 pKa = 3.79YY391 pKa = 11.84TLGSPGAAEE400 pKa = 4.16IPISVGASTPPLNVPTVDD418 pKa = 4.0GEE420 pKa = 4.61GLLTSYY426 pKa = 10.97GSLMTFSPDD435 pKa = 3.39LSTLDD440 pKa = 3.64GEE442 pKa = 4.78TIEE445 pKa = 4.49VVYY448 pKa = 10.85AGLGTTDD455 pKa = 3.0EE456 pKa = 4.55TEE458 pKa = 4.39AVDD461 pKa = 3.86LTGKK465 pKa = 9.86VALIQRR471 pKa = 11.84GSISFTEE478 pKa = 4.21KK479 pKa = 10.59SVNAQAAGAVAAIIYY494 pKa = 10.27NNEE497 pKa = 3.87PGNFGGTLGVPGDD510 pKa = 4.32YY511 pKa = 10.55IPTLSISLEE520 pKa = 4.31DD521 pKa = 3.54GTALKK526 pKa = 10.64SKK528 pKa = 10.64IEE530 pKa = 3.95ATPGYY535 pKa = 9.46EE536 pKa = 3.53ITFGIDD542 pKa = 3.1LQQDD546 pKa = 3.62LMADD550 pKa = 4.39FSSRR554 pKa = 11.84GPALPKK560 pKa = 10.65YY561 pKa = 9.93GIKK564 pKa = 9.95PDD566 pKa = 3.22ISAPGVAIRR575 pKa = 11.84SSIPAYY581 pKa = 10.35GGDD584 pKa = 3.63YY585 pKa = 10.97SEE587 pKa = 6.02AYY589 pKa = 10.4ADD591 pKa = 3.86FQGTSMATPHH601 pKa = 5.67VAGAAALLLDD611 pKa = 4.43KK612 pKa = 10.85DD613 pKa = 3.93ASLTLSEE620 pKa = 4.84IKK622 pKa = 10.92GLMTNNALKK631 pKa = 10.89LSDD634 pKa = 4.24LDD636 pKa = 3.85GNRR639 pKa = 11.84YY640 pKa = 6.73EE641 pKa = 5.52HH642 pKa = 6.98NIQGAGRR649 pKa = 11.84LDD651 pKa = 3.76LEE653 pKa = 4.53STLEE657 pKa = 4.05AKK659 pKa = 10.26AVALVQEE666 pKa = 4.34EE667 pKa = 4.63TTAVEE672 pKa = 4.47SGEE675 pKa = 4.06EE676 pKa = 4.06TPYY679 pKa = 10.17EE680 pKa = 4.21TGLLSYY686 pKa = 10.7GILDD690 pKa = 4.16GGSTSTKK697 pKa = 8.99TIEE700 pKa = 4.08VSDD703 pKa = 3.95VVGEE707 pKa = 4.14GSSYY711 pKa = 10.99SVSTEE716 pKa = 3.61WYY718 pKa = 6.5GTPPGTLSVSEE729 pKa = 4.41SSISVPVNGEE739 pKa = 3.4ASIDD743 pKa = 3.48VTIAMSNGIADD754 pKa = 3.76GNYY757 pKa = 9.38EE758 pKa = 4.1GEE760 pKa = 4.92LILTEE765 pKa = 4.55AGGHH769 pKa = 4.53QLKK772 pKa = 10.42IPVYY776 pKa = 8.6VYY778 pKa = 10.08VGEE781 pKa = 4.07PVEE784 pKa = 4.31IPVVSDD790 pKa = 3.4VAVDD794 pKa = 3.72PQFFSPNEE802 pKa = 4.03DD803 pKa = 3.1EE804 pKa = 5.8LYY806 pKa = 10.54DD807 pKa = 3.72YY808 pKa = 10.96TDD810 pKa = 3.2VYY812 pKa = 11.15FSVNAGNEE820 pKa = 4.14KK821 pKa = 10.11IALDD825 pKa = 3.46ILNPNGKK832 pKa = 8.09WLGSIDD838 pKa = 3.71EE839 pKa = 4.98SNGEE843 pKa = 4.35MEE845 pKa = 5.56PGDD848 pKa = 3.78YY849 pKa = 10.04WIQQWDD855 pKa = 3.72GTIINGPNIIKK866 pKa = 10.59LEE868 pKa = 4.35DD869 pKa = 4.31GIYY872 pKa = 9.78FASGSALINGGWIEE886 pKa = 4.35YY887 pKa = 10.34LDD889 pKa = 3.57EE890 pKa = 5.28AYY892 pKa = 10.02PFVLDD897 pKa = 3.77TAAPVSTLDD906 pKa = 3.83DD907 pKa = 3.73PQITVEE913 pKa = 4.32GDD915 pKa = 2.82VGTIRR920 pKa = 11.84GQIEE924 pKa = 4.23SDD926 pKa = 3.72LMVDD930 pKa = 3.8ALGNYY935 pKa = 9.76SGIGVEE941 pKa = 4.83AGYY944 pKa = 10.59LVDD947 pKa = 5.62GEE949 pKa = 3.9WDD951 pKa = 3.67YY952 pKa = 11.99AIGSISDD959 pKa = 3.37NGEE962 pKa = 3.56FTIEE966 pKa = 3.96VPIVEE971 pKa = 4.34GQNDD975 pKa = 4.01FEE977 pKa = 5.24VYY979 pKa = 10.52VYY981 pKa = 10.45DD982 pKa = 3.59YY983 pKa = 9.9TGNGFIEE990 pKa = 4.74PSHH993 pKa = 5.79LVSYY997 pKa = 10.35EE998 pKa = 3.89SDD1000 pKa = 3.44GEE1002 pKa = 4.36EE1003 pKa = 4.28PGEE1006 pKa = 3.99PTPGEE1011 pKa = 3.61ISLTVDD1017 pKa = 3.68PSADD1021 pKa = 2.99EE1022 pKa = 4.28VGVNEE1027 pKa = 5.11SFNLDD1032 pKa = 2.85IDD1034 pKa = 4.16FSEE1037 pKa = 5.18VEE1039 pKa = 4.58DD1040 pKa = 4.57LYY1042 pKa = 11.51SAQFSLTYY1050 pKa = 10.79DD1051 pKa = 3.46EE1052 pKa = 5.82DD1053 pKa = 4.12LVKK1056 pKa = 10.31GTTAPSVVLATYY1068 pKa = 9.55QEE1070 pKa = 4.25EE1071 pKa = 4.41QNPGVTPYY1079 pKa = 10.71VYY1081 pKa = 10.82EE1082 pKa = 4.01EE1083 pKa = 4.08TTALGNGLAKK1093 pKa = 9.51TDD1095 pKa = 3.79YY1096 pKa = 11.05VITLVGLEE1104 pKa = 3.93SGYY1107 pKa = 9.5TGEE1110 pKa = 4.61GALASFNFAGVSEE1123 pKa = 4.4GEE1125 pKa = 4.03YY1126 pKa = 10.84DD1127 pKa = 3.9FEE1129 pKa = 5.06LSNVRR1134 pKa = 11.84ALNSTSADD1142 pKa = 3.1IAVGDD1147 pKa = 3.58ITGASVTVTSDD1158 pKa = 3.59PSPGQYY1164 pKa = 10.4SITGSIDD1171 pKa = 2.93AEE1173 pKa = 4.14AFADD1177 pKa = 4.04GVDD1180 pKa = 4.61YY1181 pKa = 11.6SEE1183 pKa = 4.14TWYY1186 pKa = 10.74QGADD1190 pKa = 3.27GVHH1193 pKa = 5.53KK1194 pKa = 10.76VVVEE1198 pKa = 4.14AVDD1201 pKa = 3.88SNGSVAGVGTVRR1213 pKa = 11.84ADD1215 pKa = 3.24GTYY1218 pKa = 9.83TIEE1221 pKa = 4.28VPEE1224 pKa = 3.98GTYY1227 pKa = 7.61TVRR1230 pKa = 11.84VVVPGHH1236 pKa = 4.91ITEE1239 pKa = 4.51TAGVTVDD1246 pKa = 4.38GNEE1249 pKa = 4.08TLNFGPLTAGDD1260 pKa = 4.04VNNDD1264 pKa = 3.13GAVNLVDD1271 pKa = 4.16LQLTAKK1277 pKa = 10.33EE1278 pKa = 3.95FGKK1281 pKa = 9.22TKK1283 pKa = 10.4GSAWANAKK1291 pKa = 10.36ASAADD1296 pKa = 3.51INRR1299 pKa = 11.84DD1300 pKa = 3.54SAVDD1304 pKa = 4.13LLDD1307 pKa = 3.13ISYY1310 pKa = 10.49IIGNYY1315 pKa = 7.84EE1316 pKa = 3.63LL1317 pKa = 4.92
MM1 pKa = 6.85QSKK4 pKa = 9.78PIRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84LMSLSVVSTLFLSTVFPAVVSGAEE33 pKa = 3.7ATNRR37 pKa = 11.84KK38 pKa = 8.94FDD40 pKa = 3.94PSLFNLDD47 pKa = 3.38EE48 pKa = 4.76SKK50 pKa = 11.16LNSQMNGVTLNSSKK64 pKa = 10.66KK65 pKa = 9.4QAALNNTDD73 pKa = 2.7QSTNTGSKK81 pKa = 9.64IEE83 pKa = 5.09SYY85 pKa = 8.95TQSDD89 pKa = 3.34NSEE92 pKa = 4.1RR93 pKa = 11.84ASSVITPQNYY103 pKa = 8.99VPEE106 pKa = 4.65SDD108 pKa = 3.61STSTISVIVEE118 pKa = 3.95LSEE121 pKa = 4.2EE122 pKa = 4.53PIALYY127 pKa = 10.15EE128 pKa = 4.47SKK130 pKa = 10.71SSQNLNSGNNDD141 pKa = 2.99QRR143 pKa = 11.84SLIQKK148 pKa = 7.16QQQSFASAAKK158 pKa = 10.2RR159 pKa = 11.84LNATLKK165 pKa = 9.01TPYY168 pKa = 10.15SVVFNGYY175 pKa = 10.13AVDD178 pKa = 3.45IRR180 pKa = 11.84ADD182 pKa = 3.8RR183 pKa = 11.84VEE185 pKa = 4.36SLLAIPGVKK194 pKa = 9.52AVYY197 pKa = 9.59PDD199 pKa = 4.26SEE201 pKa = 4.48MKK203 pKa = 9.38ATPINSITPNMDD215 pKa = 2.56EE216 pKa = 4.12SAPFIGSDD224 pKa = 3.54TFWDD228 pKa = 3.09IGYY231 pKa = 9.67DD232 pKa = 3.45GAGIKK237 pKa = 10.5VGVLDD242 pKa = 3.48TGIDD246 pKa = 3.75YY247 pKa = 7.96EE248 pKa = 4.58HH249 pKa = 7.6PSLADD254 pKa = 3.69AYY256 pKa = 10.02KK257 pKa = 10.64GGYY260 pKa = 10.07DD261 pKa = 5.42FIDD264 pKa = 4.08NDD266 pKa = 4.21DD267 pKa = 4.94DD268 pKa = 4.75PYY270 pKa = 10.84EE271 pKa = 4.44TPPDD275 pKa = 4.27PNDD278 pKa = 3.55PEE280 pKa = 4.44AATDD284 pKa = 3.42HH285 pKa = 5.97GTHH288 pKa = 5.88VSGTIAGRR296 pKa = 11.84GNPLDD301 pKa = 4.36PEE303 pKa = 4.69SGTGWVRR310 pKa = 11.84GVAYY314 pKa = 10.04GADD317 pKa = 2.85IYY319 pKa = 10.95AYY321 pKa = 9.87RR322 pKa = 11.84VLGPGGSGPTSGVIAAIEE340 pKa = 3.89QSVEE344 pKa = 3.81DD345 pKa = 5.38DD346 pKa = 3.17MDD348 pKa = 5.01IINLSLGSEE357 pKa = 4.4MNNEE361 pKa = 4.06FDD363 pKa = 4.01PASVALNNAALAGVVAVVANGNSGPDD389 pKa = 3.92DD390 pKa = 3.79YY391 pKa = 11.84TLGSPGAAEE400 pKa = 4.16IPISVGASTPPLNVPTVDD418 pKa = 4.0GEE420 pKa = 4.61GLLTSYY426 pKa = 10.97GSLMTFSPDD435 pKa = 3.39LSTLDD440 pKa = 3.64GEE442 pKa = 4.78TIEE445 pKa = 4.49VVYY448 pKa = 10.85AGLGTTDD455 pKa = 3.0EE456 pKa = 4.55TEE458 pKa = 4.39AVDD461 pKa = 3.86LTGKK465 pKa = 9.86VALIQRR471 pKa = 11.84GSISFTEE478 pKa = 4.21KK479 pKa = 10.59SVNAQAAGAVAAIIYY494 pKa = 10.27NNEE497 pKa = 3.87PGNFGGTLGVPGDD510 pKa = 4.32YY511 pKa = 10.55IPTLSISLEE520 pKa = 4.31DD521 pKa = 3.54GTALKK526 pKa = 10.64SKK528 pKa = 10.64IEE530 pKa = 3.95ATPGYY535 pKa = 9.46EE536 pKa = 3.53ITFGIDD542 pKa = 3.1LQQDD546 pKa = 3.62LMADD550 pKa = 4.39FSSRR554 pKa = 11.84GPALPKK560 pKa = 10.65YY561 pKa = 9.93GIKK564 pKa = 9.95PDD566 pKa = 3.22ISAPGVAIRR575 pKa = 11.84SSIPAYY581 pKa = 10.35GGDD584 pKa = 3.63YY585 pKa = 10.97SEE587 pKa = 6.02AYY589 pKa = 10.4ADD591 pKa = 3.86FQGTSMATPHH601 pKa = 5.67VAGAAALLLDD611 pKa = 4.43KK612 pKa = 10.85DD613 pKa = 3.93ASLTLSEE620 pKa = 4.84IKK622 pKa = 10.92GLMTNNALKK631 pKa = 10.89LSDD634 pKa = 4.24LDD636 pKa = 3.85GNRR639 pKa = 11.84YY640 pKa = 6.73EE641 pKa = 5.52HH642 pKa = 6.98NIQGAGRR649 pKa = 11.84LDD651 pKa = 3.76LEE653 pKa = 4.53STLEE657 pKa = 4.05AKK659 pKa = 10.26AVALVQEE666 pKa = 4.34EE667 pKa = 4.63TTAVEE672 pKa = 4.47SGEE675 pKa = 4.06EE676 pKa = 4.06TPYY679 pKa = 10.17EE680 pKa = 4.21TGLLSYY686 pKa = 10.7GILDD690 pKa = 4.16GGSTSTKK697 pKa = 8.99TIEE700 pKa = 4.08VSDD703 pKa = 3.95VVGEE707 pKa = 4.14GSSYY711 pKa = 10.99SVSTEE716 pKa = 3.61WYY718 pKa = 6.5GTPPGTLSVSEE729 pKa = 4.41SSISVPVNGEE739 pKa = 3.4ASIDD743 pKa = 3.48VTIAMSNGIADD754 pKa = 3.76GNYY757 pKa = 9.38EE758 pKa = 4.1GEE760 pKa = 4.92LILTEE765 pKa = 4.55AGGHH769 pKa = 4.53QLKK772 pKa = 10.42IPVYY776 pKa = 8.6VYY778 pKa = 10.08VGEE781 pKa = 4.07PVEE784 pKa = 4.31IPVVSDD790 pKa = 3.4VAVDD794 pKa = 3.72PQFFSPNEE802 pKa = 4.03DD803 pKa = 3.1EE804 pKa = 5.8LYY806 pKa = 10.54DD807 pKa = 3.72YY808 pKa = 10.96TDD810 pKa = 3.2VYY812 pKa = 11.15FSVNAGNEE820 pKa = 4.14KK821 pKa = 10.11IALDD825 pKa = 3.46ILNPNGKK832 pKa = 8.09WLGSIDD838 pKa = 3.71EE839 pKa = 4.98SNGEE843 pKa = 4.35MEE845 pKa = 5.56PGDD848 pKa = 3.78YY849 pKa = 10.04WIQQWDD855 pKa = 3.72GTIINGPNIIKK866 pKa = 10.59LEE868 pKa = 4.35DD869 pKa = 4.31GIYY872 pKa = 9.78FASGSALINGGWIEE886 pKa = 4.35YY887 pKa = 10.34LDD889 pKa = 3.57EE890 pKa = 5.28AYY892 pKa = 10.02PFVLDD897 pKa = 3.77TAAPVSTLDD906 pKa = 3.83DD907 pKa = 3.73PQITVEE913 pKa = 4.32GDD915 pKa = 2.82VGTIRR920 pKa = 11.84GQIEE924 pKa = 4.23SDD926 pKa = 3.72LMVDD930 pKa = 3.8ALGNYY935 pKa = 9.76SGIGVEE941 pKa = 4.83AGYY944 pKa = 10.59LVDD947 pKa = 5.62GEE949 pKa = 3.9WDD951 pKa = 3.67YY952 pKa = 11.99AIGSISDD959 pKa = 3.37NGEE962 pKa = 3.56FTIEE966 pKa = 3.96VPIVEE971 pKa = 4.34GQNDD975 pKa = 4.01FEE977 pKa = 5.24VYY979 pKa = 10.52VYY981 pKa = 10.45DD982 pKa = 3.59YY983 pKa = 9.9TGNGFIEE990 pKa = 4.74PSHH993 pKa = 5.79LVSYY997 pKa = 10.35EE998 pKa = 3.89SDD1000 pKa = 3.44GEE1002 pKa = 4.36EE1003 pKa = 4.28PGEE1006 pKa = 3.99PTPGEE1011 pKa = 3.61ISLTVDD1017 pKa = 3.68PSADD1021 pKa = 2.99EE1022 pKa = 4.28VGVNEE1027 pKa = 5.11SFNLDD1032 pKa = 2.85IDD1034 pKa = 4.16FSEE1037 pKa = 5.18VEE1039 pKa = 4.58DD1040 pKa = 4.57LYY1042 pKa = 11.51SAQFSLTYY1050 pKa = 10.79DD1051 pKa = 3.46EE1052 pKa = 5.82DD1053 pKa = 4.12LVKK1056 pKa = 10.31GTTAPSVVLATYY1068 pKa = 9.55QEE1070 pKa = 4.25EE1071 pKa = 4.41QNPGVTPYY1079 pKa = 10.71VYY1081 pKa = 10.82EE1082 pKa = 4.01EE1083 pKa = 4.08TTALGNGLAKK1093 pKa = 9.51TDD1095 pKa = 3.79YY1096 pKa = 11.05VITLVGLEE1104 pKa = 3.93SGYY1107 pKa = 9.5TGEE1110 pKa = 4.61GALASFNFAGVSEE1123 pKa = 4.4GEE1125 pKa = 4.03YY1126 pKa = 10.84DD1127 pKa = 3.9FEE1129 pKa = 5.06LSNVRR1134 pKa = 11.84ALNSTSADD1142 pKa = 3.1IAVGDD1147 pKa = 3.58ITGASVTVTSDD1158 pKa = 3.59PSPGQYY1164 pKa = 10.4SITGSIDD1171 pKa = 2.93AEE1173 pKa = 4.14AFADD1177 pKa = 4.04GVDD1180 pKa = 4.61YY1181 pKa = 11.6SEE1183 pKa = 4.14TWYY1186 pKa = 10.74QGADD1190 pKa = 3.27GVHH1193 pKa = 5.53KK1194 pKa = 10.76VVVEE1198 pKa = 4.14AVDD1201 pKa = 3.88SNGSVAGVGTVRR1213 pKa = 11.84ADD1215 pKa = 3.24GTYY1218 pKa = 9.83TIEE1221 pKa = 4.28VPEE1224 pKa = 3.98GTYY1227 pKa = 7.61TVRR1230 pKa = 11.84VVVPGHH1236 pKa = 4.91ITEE1239 pKa = 4.51TAGVTVDD1246 pKa = 4.38GNEE1249 pKa = 4.08TLNFGPLTAGDD1260 pKa = 4.04VNNDD1264 pKa = 3.13GAVNLVDD1271 pKa = 4.16LQLTAKK1277 pKa = 10.33EE1278 pKa = 3.95FGKK1281 pKa = 9.22TKK1283 pKa = 10.4GSAWANAKK1291 pKa = 10.36ASAADD1296 pKa = 3.51INRR1299 pKa = 11.84DD1300 pKa = 3.54SAVDD1304 pKa = 4.13LLDD1307 pKa = 3.13ISYY1310 pKa = 10.49IIGNYY1315 pKa = 7.84EE1316 pKa = 3.63LL1317 pKa = 4.92
Molecular weight: 138.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A069DFT5|A0A069DFT5_9BACL LytR_cpsA_psr domain-containing protein OS=Paenibacillus sp. TCA20 OX=1499968 GN=TCA2_3593 PE=3 SV=1
MM1 pKa = 7.82RR2 pKa = 11.84PTFRR6 pKa = 11.84PNVSKK11 pKa = 10.72RR12 pKa = 11.84KK13 pKa = 8.96KK14 pKa = 8.25VHH16 pKa = 5.49GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.87VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.79VLSAA44 pKa = 4.05
MM1 pKa = 7.82RR2 pKa = 11.84PTFRR6 pKa = 11.84PNVSKK11 pKa = 10.72RR12 pKa = 11.84KK13 pKa = 8.96KK14 pKa = 8.25VHH16 pKa = 5.49GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.87VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.79VLSAA44 pKa = 4.05
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1621612 |
39 |
4464 |
310.3 |
34.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.689 ± 0.032 | 0.685 ± 0.009 |
5.269 ± 0.027 | 7.318 ± 0.04 |
3.977 ± 0.022 | 7.203 ± 0.035 |
2.214 ± 0.019 | 7.034 ± 0.033 |
5.296 ± 0.029 | 9.774 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.987 ± 0.016 | 4.011 ± 0.025 |
3.916 ± 0.024 | 3.964 ± 0.022 |
4.727 ± 0.03 | 6.527 ± 0.029 |
5.533 ± 0.035 | 6.945 ± 0.029 |
1.194 ± 0.016 | 3.737 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
