
Magnaporthe oryzae chrysovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Chrysoviridae; Chrysovirus; unclassified Chrysovirus
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
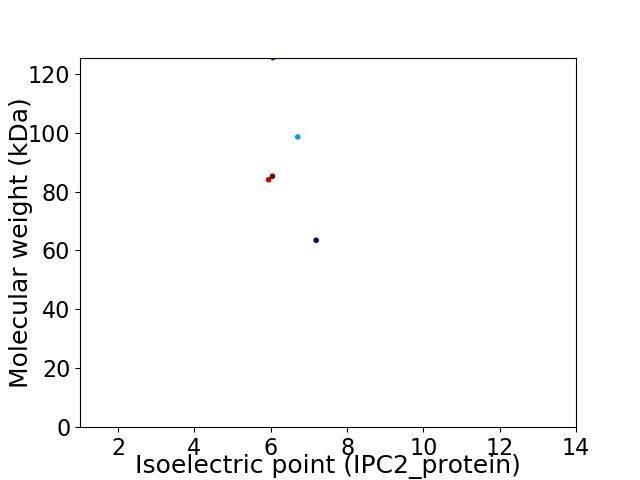
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
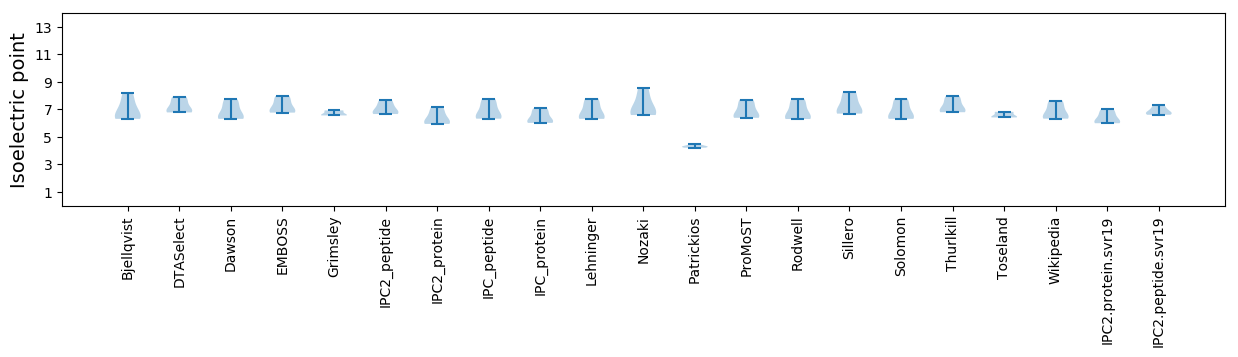
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E0D6V6|E0D6V6_9VIRU Uncharacterized protein OS=Magnaporthe oryzae chrysovirus 1 OX=764348 PE=4 SV=1
MM1 pKa = 7.64GLTLDD6 pKa = 4.16PAHH9 pKa = 7.39RR10 pKa = 11.84WRR12 pKa = 11.84STDD15 pKa = 3.5LAFAPVNEE23 pKa = 4.36VSVQSAVRR31 pKa = 11.84GRR33 pKa = 11.84DD34 pKa = 3.35DD35 pKa = 4.24SEE37 pKa = 4.15AAGTGFTGDD46 pKa = 3.49KK47 pKa = 9.3ATKK50 pKa = 9.54AWYY53 pKa = 9.64DD54 pKa = 3.25GAGRR58 pKa = 11.84VPIDD62 pKa = 4.63DD63 pKa = 3.84MCLAAGMRR71 pKa = 11.84AGVLRR76 pKa = 11.84LAVEE80 pKa = 4.44VGSARR85 pKa = 11.84PSSADD90 pKa = 3.09EE91 pKa = 4.95SVVQWRR97 pKa = 11.84SVPYY101 pKa = 10.12SYY103 pKa = 10.85VGKK106 pKa = 9.06PLTVSLSHH114 pKa = 7.21AGRR117 pKa = 11.84HH118 pKa = 4.62FVARR122 pKa = 11.84PALNEE127 pKa = 3.91NVAMAMYY134 pKa = 8.0EE135 pKa = 4.2APTADD140 pKa = 2.93KK141 pKa = 10.21WVAATNFKK149 pKa = 10.66LPRR152 pKa = 11.84TVAAPGAAPQVPGLPNGGGGANLGLPNNFDD182 pKa = 3.12AVRR185 pKa = 11.84RR186 pKa = 11.84VLVEE190 pKa = 3.67CARR193 pKa = 11.84GDD195 pKa = 3.55DD196 pKa = 3.38YY197 pKa = 11.65GYY199 pKa = 11.24RR200 pKa = 11.84LFSMARR206 pKa = 11.84VVLHH210 pKa = 6.77AEE212 pKa = 3.91TMRR215 pKa = 11.84RR216 pKa = 11.84SGISPRR222 pKa = 11.84ATPSMDD228 pKa = 3.56DD229 pKa = 3.18QNMFSITTGDD239 pKa = 3.72TPHH242 pKa = 6.5LTEE245 pKa = 4.48AQINNYY251 pKa = 9.7AYY253 pKa = 10.17AYY255 pKa = 10.04NHH257 pKa = 6.41TEE259 pKa = 3.34QSPQYY264 pKa = 10.13RR265 pKa = 11.84AFLAMGLRR273 pKa = 11.84GVGHH277 pKa = 5.73YY278 pKa = 9.71AIPGTIYY285 pKa = 10.8SDD287 pKa = 2.78GDD289 pKa = 3.98YY290 pKa = 10.87PVEE293 pKa = 4.22CAVNHH298 pKa = 6.7PIAFVRR304 pKa = 11.84VGGPPPANVAPDD316 pKa = 3.88PAHH319 pKa = 5.06YY320 pKa = 8.69TAVLSNPGLALSYY333 pKa = 9.15YY334 pKa = 8.13WAYY337 pKa = 10.18AYY339 pKa = 11.39SMGLGRR345 pKa = 11.84VAGAILAQASIAPHH359 pKa = 5.54IWGSAAVAPYY369 pKa = 10.38KK370 pKa = 10.89NCTPKK375 pKa = 10.63LDD377 pKa = 3.55AAAYY381 pKa = 10.23LLLPDD386 pKa = 4.17QEE388 pKa = 4.75TAHH391 pKa = 5.55VTADD395 pKa = 3.31SARR398 pKa = 11.84EE399 pKa = 3.96LVANAAVLSEE409 pKa = 4.8AYY411 pKa = 10.15LAGIGATLLSARR423 pKa = 11.84DD424 pKa = 3.69SGHH427 pKa = 6.42QDD429 pKa = 2.68TALMMRR435 pKa = 11.84AVTEE439 pKa = 4.1KK440 pKa = 10.88LSDD443 pKa = 3.43PEE445 pKa = 4.17TRR447 pKa = 11.84RR448 pKa = 11.84GAMLSITSRR457 pKa = 11.84LCPGGVGMEE466 pKa = 4.07WLSPFSYY473 pKa = 10.7DD474 pKa = 4.02VLDD477 pKa = 3.75GTEE480 pKa = 3.63RR481 pKa = 11.84CIRR484 pKa = 11.84AWRR487 pKa = 11.84NHH489 pKa = 4.84GFLLALYY496 pKa = 7.47DD497 pKa = 4.03TSPVAALAPLFSTGVPMNNSLLDD520 pKa = 3.64KK521 pKa = 10.73KK522 pKa = 11.04SVVTGAEE529 pKa = 4.05YY530 pKa = 9.47PQLVACALAGRR541 pKa = 11.84AEE543 pKa = 3.93LAGRR547 pKa = 11.84CEE549 pKa = 4.39RR550 pKa = 11.84PSPAYY555 pKa = 10.0LAALAGHH562 pKa = 6.14SARR565 pKa = 11.84MRR567 pKa = 11.84AWTVVVTVLGVVPPASDD584 pKa = 4.64DD585 pKa = 3.57EE586 pKa = 4.45DD587 pKa = 3.96HH588 pKa = 7.87ADD590 pKa = 3.64VQEE593 pKa = 4.28QAASRR598 pKa = 11.84QEE600 pKa = 4.07SSSSVRR606 pKa = 11.84PLSSQSDD613 pKa = 3.44RR614 pKa = 11.84SVTRR618 pKa = 11.84GHH620 pKa = 6.0GTAEE624 pKa = 4.08PAQAGAGPSSPPPVAPLRR642 pKa = 11.84GMRR645 pKa = 11.84LGSRR649 pKa = 11.84ARR651 pKa = 11.84SHH653 pKa = 6.46KK654 pKa = 10.44GSLSAPQDD662 pKa = 4.46PIPEE666 pKa = 4.29TGEE669 pKa = 3.95EE670 pKa = 4.38PVRR673 pKa = 11.84QPDD676 pKa = 3.64GAGAKK681 pKa = 9.85PKK683 pKa = 10.32VQAILEE689 pKa = 4.31APKK692 pKa = 9.98PALEE696 pKa = 4.28PPSWNSWASEE706 pKa = 4.21VASVEE711 pKa = 3.82ARR713 pKa = 11.84LAGADD718 pKa = 3.34EE719 pKa = 4.67GPRR722 pKa = 11.84GKK724 pKa = 9.61IVEE727 pKa = 4.01PEE729 pKa = 3.82YY730 pKa = 10.93RR731 pKa = 11.84GIAPSRR737 pKa = 11.84STTTVATSRR746 pKa = 11.84SVVPSAASAASGTVVSMGRR765 pKa = 11.84RR766 pKa = 11.84AKK768 pKa = 9.97GKK770 pKa = 8.31EE771 pKa = 3.96RR772 pKa = 11.84EE773 pKa = 4.17VEE775 pKa = 3.78RR776 pKa = 11.84ADD778 pKa = 3.58QVEE781 pKa = 4.41SSSGSSDD788 pKa = 3.28QASGGSRR795 pKa = 11.84GEE797 pKa = 4.04EE798 pKa = 3.84LL799 pKa = 4.08
MM1 pKa = 7.64GLTLDD6 pKa = 4.16PAHH9 pKa = 7.39RR10 pKa = 11.84WRR12 pKa = 11.84STDD15 pKa = 3.5LAFAPVNEE23 pKa = 4.36VSVQSAVRR31 pKa = 11.84GRR33 pKa = 11.84DD34 pKa = 3.35DD35 pKa = 4.24SEE37 pKa = 4.15AAGTGFTGDD46 pKa = 3.49KK47 pKa = 9.3ATKK50 pKa = 9.54AWYY53 pKa = 9.64DD54 pKa = 3.25GAGRR58 pKa = 11.84VPIDD62 pKa = 4.63DD63 pKa = 3.84MCLAAGMRR71 pKa = 11.84AGVLRR76 pKa = 11.84LAVEE80 pKa = 4.44VGSARR85 pKa = 11.84PSSADD90 pKa = 3.09EE91 pKa = 4.95SVVQWRR97 pKa = 11.84SVPYY101 pKa = 10.12SYY103 pKa = 10.85VGKK106 pKa = 9.06PLTVSLSHH114 pKa = 7.21AGRR117 pKa = 11.84HH118 pKa = 4.62FVARR122 pKa = 11.84PALNEE127 pKa = 3.91NVAMAMYY134 pKa = 8.0EE135 pKa = 4.2APTADD140 pKa = 2.93KK141 pKa = 10.21WVAATNFKK149 pKa = 10.66LPRR152 pKa = 11.84TVAAPGAAPQVPGLPNGGGGANLGLPNNFDD182 pKa = 3.12AVRR185 pKa = 11.84RR186 pKa = 11.84VLVEE190 pKa = 3.67CARR193 pKa = 11.84GDD195 pKa = 3.55DD196 pKa = 3.38YY197 pKa = 11.65GYY199 pKa = 11.24RR200 pKa = 11.84LFSMARR206 pKa = 11.84VVLHH210 pKa = 6.77AEE212 pKa = 3.91TMRR215 pKa = 11.84RR216 pKa = 11.84SGISPRR222 pKa = 11.84ATPSMDD228 pKa = 3.56DD229 pKa = 3.18QNMFSITTGDD239 pKa = 3.72TPHH242 pKa = 6.5LTEE245 pKa = 4.48AQINNYY251 pKa = 9.7AYY253 pKa = 10.17AYY255 pKa = 10.04NHH257 pKa = 6.41TEE259 pKa = 3.34QSPQYY264 pKa = 10.13RR265 pKa = 11.84AFLAMGLRR273 pKa = 11.84GVGHH277 pKa = 5.73YY278 pKa = 9.71AIPGTIYY285 pKa = 10.8SDD287 pKa = 2.78GDD289 pKa = 3.98YY290 pKa = 10.87PVEE293 pKa = 4.22CAVNHH298 pKa = 6.7PIAFVRR304 pKa = 11.84VGGPPPANVAPDD316 pKa = 3.88PAHH319 pKa = 5.06YY320 pKa = 8.69TAVLSNPGLALSYY333 pKa = 9.15YY334 pKa = 8.13WAYY337 pKa = 10.18AYY339 pKa = 11.39SMGLGRR345 pKa = 11.84VAGAILAQASIAPHH359 pKa = 5.54IWGSAAVAPYY369 pKa = 10.38KK370 pKa = 10.89NCTPKK375 pKa = 10.63LDD377 pKa = 3.55AAAYY381 pKa = 10.23LLLPDD386 pKa = 4.17QEE388 pKa = 4.75TAHH391 pKa = 5.55VTADD395 pKa = 3.31SARR398 pKa = 11.84EE399 pKa = 3.96LVANAAVLSEE409 pKa = 4.8AYY411 pKa = 10.15LAGIGATLLSARR423 pKa = 11.84DD424 pKa = 3.69SGHH427 pKa = 6.42QDD429 pKa = 2.68TALMMRR435 pKa = 11.84AVTEE439 pKa = 4.1KK440 pKa = 10.88LSDD443 pKa = 3.43PEE445 pKa = 4.17TRR447 pKa = 11.84RR448 pKa = 11.84GAMLSITSRR457 pKa = 11.84LCPGGVGMEE466 pKa = 4.07WLSPFSYY473 pKa = 10.7DD474 pKa = 4.02VLDD477 pKa = 3.75GTEE480 pKa = 3.63RR481 pKa = 11.84CIRR484 pKa = 11.84AWRR487 pKa = 11.84NHH489 pKa = 4.84GFLLALYY496 pKa = 7.47DD497 pKa = 4.03TSPVAALAPLFSTGVPMNNSLLDD520 pKa = 3.64KK521 pKa = 10.73KK522 pKa = 11.04SVVTGAEE529 pKa = 4.05YY530 pKa = 9.47PQLVACALAGRR541 pKa = 11.84AEE543 pKa = 3.93LAGRR547 pKa = 11.84CEE549 pKa = 4.39RR550 pKa = 11.84PSPAYY555 pKa = 10.0LAALAGHH562 pKa = 6.14SARR565 pKa = 11.84MRR567 pKa = 11.84AWTVVVTVLGVVPPASDD584 pKa = 4.64DD585 pKa = 3.57EE586 pKa = 4.45DD587 pKa = 3.96HH588 pKa = 7.87ADD590 pKa = 3.64VQEE593 pKa = 4.28QAASRR598 pKa = 11.84QEE600 pKa = 4.07SSSSVRR606 pKa = 11.84PLSSQSDD613 pKa = 3.44RR614 pKa = 11.84SVTRR618 pKa = 11.84GHH620 pKa = 6.0GTAEE624 pKa = 4.08PAQAGAGPSSPPPVAPLRR642 pKa = 11.84GMRR645 pKa = 11.84LGSRR649 pKa = 11.84ARR651 pKa = 11.84SHH653 pKa = 6.46KK654 pKa = 10.44GSLSAPQDD662 pKa = 4.46PIPEE666 pKa = 4.29TGEE669 pKa = 3.95EE670 pKa = 4.38PVRR673 pKa = 11.84QPDD676 pKa = 3.64GAGAKK681 pKa = 9.85PKK683 pKa = 10.32VQAILEE689 pKa = 4.31APKK692 pKa = 9.98PALEE696 pKa = 4.28PPSWNSWASEE706 pKa = 4.21VASVEE711 pKa = 3.82ARR713 pKa = 11.84LAGADD718 pKa = 3.34EE719 pKa = 4.67GPRR722 pKa = 11.84GKK724 pKa = 9.61IVEE727 pKa = 4.01PEE729 pKa = 3.82YY730 pKa = 10.93RR731 pKa = 11.84GIAPSRR737 pKa = 11.84STTTVATSRR746 pKa = 11.84SVVPSAASAASGTVVSMGRR765 pKa = 11.84RR766 pKa = 11.84AKK768 pKa = 9.97GKK770 pKa = 8.31EE771 pKa = 3.96RR772 pKa = 11.84EE773 pKa = 4.17VEE775 pKa = 3.78RR776 pKa = 11.84ADD778 pKa = 3.58QVEE781 pKa = 4.41SSSGSSDD788 pKa = 3.28QASGGSRR795 pKa = 11.84GEE797 pKa = 4.04EE798 pKa = 3.84LL799 pKa = 4.08
Molecular weight: 84.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I7H3R3|I7H3R3_9VIRU Uncharacterized protein OS=Magnaporthe oryzae chrysovirus 1 OX=764348 PE=4 SV=1
MM1 pKa = 7.99DD2 pKa = 5.73AKK4 pKa = 10.81QSGAGARR11 pKa = 11.84VSGWWPDD18 pKa = 3.09RR19 pKa = 11.84QVIGRR24 pKa = 11.84EE25 pKa = 3.91ALRR28 pKa = 11.84QRR30 pKa = 11.84TSRR33 pKa = 11.84TLGAMGGAVGVSDD46 pKa = 3.88TPRR49 pKa = 11.84SFFGGGEE56 pKa = 4.18VPTEE60 pKa = 3.77RR61 pKa = 11.84LRR63 pKa = 11.84RR64 pKa = 11.84GLPSEE69 pKa = 4.09VAGMFTAEE77 pKa = 4.45QLSEE81 pKa = 4.21MLDD84 pKa = 3.3MSAEE88 pKa = 4.0QRR90 pKa = 11.84VKK92 pKa = 10.97ALADD96 pKa = 2.89AWEE99 pKa = 4.21RR100 pKa = 11.84LAAAEE105 pKa = 4.37ASTGPTSSGPQTPARR120 pKa = 11.84RR121 pKa = 11.84APRR124 pKa = 11.84SVSDD128 pKa = 3.64YY129 pKa = 10.65NTTQWVAAAANDD141 pKa = 5.24GIRR144 pKa = 11.84VTPTLTAAARR154 pKa = 11.84AVAPALNRR162 pKa = 11.84AVVPGGCQPHH172 pKa = 6.68ASYY175 pKa = 11.28GRR177 pKa = 11.84ATVATGTGDD186 pKa = 3.26IAPPSARR193 pKa = 11.84QTAVVQAIGEE203 pKa = 4.47SVPTEE208 pKa = 3.88PVDD211 pKa = 4.28AEE213 pKa = 4.5VKK215 pKa = 10.53EE216 pKa = 4.65GTPSPAAPPRR226 pKa = 11.84LLCGTATLLALPQVCCGSALDD247 pKa = 4.58HH248 pKa = 6.55GCAAAPCAVGQVSGAAAAVALASRR272 pKa = 11.84PVVEE276 pKa = 4.87PDD278 pKa = 3.23ADD280 pKa = 3.54ASSMGEE286 pKa = 3.52RR287 pKa = 11.84WKK289 pKa = 11.33VMSTLAARR297 pKa = 11.84LGPIAQCHH305 pKa = 5.74FGSCRR310 pKa = 11.84DD311 pKa = 3.41VCACTHH317 pKa = 6.11RR318 pKa = 11.84HH319 pKa = 5.09PLARR323 pKa = 11.84ALMGIGVGTEE333 pKa = 3.86EE334 pKa = 3.77LRR336 pKa = 11.84GARR339 pKa = 11.84VHH341 pKa = 6.32LTTPQEE347 pKa = 4.27SRR349 pKa = 11.84SVASVLLEE357 pKa = 3.87KK358 pKa = 10.92VGGGSRR364 pKa = 11.84VEE366 pKa = 3.83RR367 pKa = 11.84LAVEE371 pKa = 4.36FVKK374 pKa = 10.95AVDD377 pKa = 4.28GPVLTSLVVASRR389 pKa = 11.84DD390 pKa = 3.48GKK392 pKa = 10.93ADD394 pKa = 3.54NLLTLVAEE402 pKa = 4.86AMAQGSLAAPRR413 pKa = 11.84TMVTFGRR420 pKa = 11.84GDD422 pKa = 3.26GGGPAFDD429 pKa = 4.46PNKK432 pKa = 10.74LYY434 pKa = 10.46DD435 pKa = 3.44ACVYY439 pKa = 10.53SGEE442 pKa = 4.34RR443 pKa = 11.84GHH445 pKa = 7.02KK446 pKa = 10.37LGVLSRR452 pKa = 11.84AVWLEE457 pKa = 3.55SDD459 pKa = 5.11ADD461 pKa = 4.17VHH463 pKa = 6.53WMPHH467 pKa = 5.24SNSLRR472 pKa = 11.84VTLQCWNRR480 pKa = 11.84RR481 pKa = 11.84GDD483 pKa = 3.94CKK485 pKa = 10.81GVSAVIGPTTWRR497 pKa = 11.84PHH499 pKa = 5.75VGNSSWEE506 pKa = 4.08TLGSDD511 pKa = 4.89VYY513 pKa = 11.08RR514 pKa = 11.84DD515 pKa = 3.23WFGLVVARR523 pKa = 11.84RR524 pKa = 11.84GGGDD528 pKa = 3.03TTVRR532 pKa = 11.84HH533 pKa = 5.13QLSYY537 pKa = 10.86NPGMEE542 pKa = 4.09ADD544 pKa = 3.55QLGGLLAHH552 pKa = 6.91SGCGVYY558 pKa = 10.64VSGADD563 pKa = 3.79DD564 pKa = 4.17AATVAAVHH572 pKa = 6.62PSLQVTYY579 pKa = 7.87THH581 pKa = 6.6ITLGSPTASKK591 pKa = 10.57LVQAATALDD600 pKa = 3.65EE601 pKa = 4.32FVRR604 pKa = 11.84RR605 pKa = 11.84GGAHH609 pKa = 6.43RR610 pKa = 11.84PP611 pKa = 3.54
MM1 pKa = 7.99DD2 pKa = 5.73AKK4 pKa = 10.81QSGAGARR11 pKa = 11.84VSGWWPDD18 pKa = 3.09RR19 pKa = 11.84QVIGRR24 pKa = 11.84EE25 pKa = 3.91ALRR28 pKa = 11.84QRR30 pKa = 11.84TSRR33 pKa = 11.84TLGAMGGAVGVSDD46 pKa = 3.88TPRR49 pKa = 11.84SFFGGGEE56 pKa = 4.18VPTEE60 pKa = 3.77RR61 pKa = 11.84LRR63 pKa = 11.84RR64 pKa = 11.84GLPSEE69 pKa = 4.09VAGMFTAEE77 pKa = 4.45QLSEE81 pKa = 4.21MLDD84 pKa = 3.3MSAEE88 pKa = 4.0QRR90 pKa = 11.84VKK92 pKa = 10.97ALADD96 pKa = 2.89AWEE99 pKa = 4.21RR100 pKa = 11.84LAAAEE105 pKa = 4.37ASTGPTSSGPQTPARR120 pKa = 11.84RR121 pKa = 11.84APRR124 pKa = 11.84SVSDD128 pKa = 3.64YY129 pKa = 10.65NTTQWVAAAANDD141 pKa = 5.24GIRR144 pKa = 11.84VTPTLTAAARR154 pKa = 11.84AVAPALNRR162 pKa = 11.84AVVPGGCQPHH172 pKa = 6.68ASYY175 pKa = 11.28GRR177 pKa = 11.84ATVATGTGDD186 pKa = 3.26IAPPSARR193 pKa = 11.84QTAVVQAIGEE203 pKa = 4.47SVPTEE208 pKa = 3.88PVDD211 pKa = 4.28AEE213 pKa = 4.5VKK215 pKa = 10.53EE216 pKa = 4.65GTPSPAAPPRR226 pKa = 11.84LLCGTATLLALPQVCCGSALDD247 pKa = 4.58HH248 pKa = 6.55GCAAAPCAVGQVSGAAAAVALASRR272 pKa = 11.84PVVEE276 pKa = 4.87PDD278 pKa = 3.23ADD280 pKa = 3.54ASSMGEE286 pKa = 3.52RR287 pKa = 11.84WKK289 pKa = 11.33VMSTLAARR297 pKa = 11.84LGPIAQCHH305 pKa = 5.74FGSCRR310 pKa = 11.84DD311 pKa = 3.41VCACTHH317 pKa = 6.11RR318 pKa = 11.84HH319 pKa = 5.09PLARR323 pKa = 11.84ALMGIGVGTEE333 pKa = 3.86EE334 pKa = 3.77LRR336 pKa = 11.84GARR339 pKa = 11.84VHH341 pKa = 6.32LTTPQEE347 pKa = 4.27SRR349 pKa = 11.84SVASVLLEE357 pKa = 3.87KK358 pKa = 10.92VGGGSRR364 pKa = 11.84VEE366 pKa = 3.83RR367 pKa = 11.84LAVEE371 pKa = 4.36FVKK374 pKa = 10.95AVDD377 pKa = 4.28GPVLTSLVVASRR389 pKa = 11.84DD390 pKa = 3.48GKK392 pKa = 10.93ADD394 pKa = 3.54NLLTLVAEE402 pKa = 4.86AMAQGSLAAPRR413 pKa = 11.84TMVTFGRR420 pKa = 11.84GDD422 pKa = 3.26GGGPAFDD429 pKa = 4.46PNKK432 pKa = 10.74LYY434 pKa = 10.46DD435 pKa = 3.44ACVYY439 pKa = 10.53SGEE442 pKa = 4.34RR443 pKa = 11.84GHH445 pKa = 7.02KK446 pKa = 10.37LGVLSRR452 pKa = 11.84AVWLEE457 pKa = 3.55SDD459 pKa = 5.11ADD461 pKa = 4.17VHH463 pKa = 6.53WMPHH467 pKa = 5.24SNSLRR472 pKa = 11.84VTLQCWNRR480 pKa = 11.84RR481 pKa = 11.84GDD483 pKa = 3.94CKK485 pKa = 10.81GVSAVIGPTTWRR497 pKa = 11.84PHH499 pKa = 5.75VGNSSWEE506 pKa = 4.08TLGSDD511 pKa = 4.89VYY513 pKa = 11.08RR514 pKa = 11.84DD515 pKa = 3.23WFGLVVARR523 pKa = 11.84RR524 pKa = 11.84GGGDD528 pKa = 3.03TTVRR532 pKa = 11.84HH533 pKa = 5.13QLSYY537 pKa = 10.86NPGMEE542 pKa = 4.09ADD544 pKa = 3.55QLGGLLAHH552 pKa = 6.91SGCGVYY558 pKa = 10.64VSGADD563 pKa = 3.79DD564 pKa = 4.17AATVAAVHH572 pKa = 6.62PSLQVTYY579 pKa = 7.87THH581 pKa = 6.6ITLGSPTASKK591 pKa = 10.57LVQAATALDD600 pKa = 3.65EE601 pKa = 4.32FVRR604 pKa = 11.84RR605 pKa = 11.84GGAHH609 pKa = 6.43RR610 pKa = 11.84PP611 pKa = 3.54
Molecular weight: 63.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4283 |
611 |
1127 |
856.6 |
91.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.565 ± 0.417 | 1.378 ± 0.154 |
5.463 ± 0.25 | 5.09 ± 0.435 |
1.915 ± 0.365 | 9.339 ± 0.721 |
2.241 ± 0.055 | 2.241 ± 0.277 |
2.405 ± 0.486 | 8.686 ± 0.605 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.358 ± 0.214 | 2.405 ± 0.172 |
5.907 ± 0.642 | 3.222 ± 0.172 |
7.798 ± 0.151 | 7.635 ± 0.488 |
5.347 ± 0.51 | 8.545 ± 0.283 |
1.471 ± 0.173 | 2.989 ± 0.371 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
