
Sathuvachari virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus; unclassified Orbivirus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
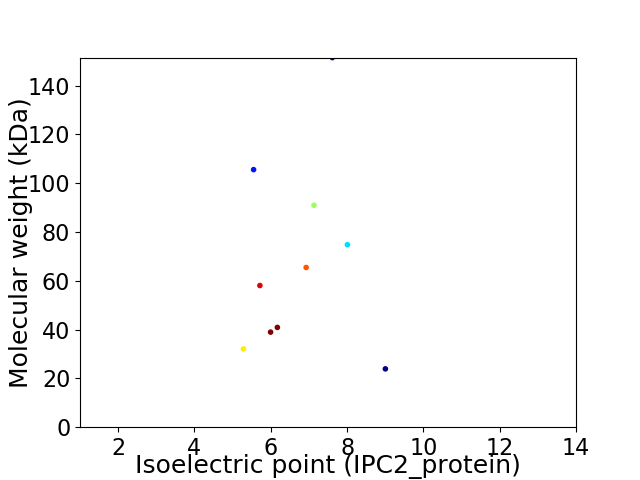
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M1H1Z8|M1H1Z8_9REOV Outer capsid protein VP5 OS=Sathuvachari virus OX=1289939 PE=3 SV=1
MM1 pKa = 7.37TRR3 pKa = 11.84LTLLAPGDD11 pKa = 4.07VIRR14 pKa = 11.84KK15 pKa = 9.61AEE17 pKa = 3.98QEE19 pKa = 4.01LKK21 pKa = 10.7ARR23 pKa = 11.84RR24 pKa = 11.84IDD26 pKa = 3.87VLIQNDD32 pKa = 3.65AEE34 pKa = 4.34EE35 pKa = 4.73GGKK38 pKa = 8.31EE39 pKa = 3.97QEE41 pKa = 4.13RR42 pKa = 11.84KK43 pKa = 10.36EE44 pKa = 3.82EE45 pKa = 4.24DD46 pKa = 3.16EE47 pKa = 4.75AGGKK51 pKa = 9.09RR52 pKa = 11.84VQEE55 pKa = 4.14STNEE59 pKa = 3.82RR60 pKa = 11.84KK61 pKa = 8.13EE62 pKa = 4.16TGNATEE68 pKa = 4.66RR69 pKa = 11.84GGEE72 pKa = 4.08GTQKK76 pKa = 10.62NDD78 pKa = 2.86EE79 pKa = 4.91DD80 pKa = 3.78GGEE83 pKa = 4.1RR84 pKa = 11.84QGRR87 pKa = 11.84EE88 pKa = 3.83AEE90 pKa = 3.92EE91 pKa = 4.1VQIHH95 pKa = 6.16KK96 pKa = 10.7AFSEE100 pKa = 4.19NKK102 pKa = 9.85GAGDD106 pKa = 4.32RR107 pKa = 11.84DD108 pKa = 4.24DD109 pKa = 5.04EE110 pKa = 4.51VQATKK115 pKa = 9.69QEE117 pKa = 4.12KK118 pKa = 9.94EE119 pKa = 4.12DD120 pKa = 3.75MVVLDD125 pKa = 4.61SEE127 pKa = 4.57VQKK130 pKa = 11.37VIDD133 pKa = 3.63KK134 pKa = 10.9LYY136 pKa = 10.69GIRR139 pKa = 11.84LQTIDD144 pKa = 4.41TITEE148 pKa = 3.54IGDD151 pKa = 3.68RR152 pKa = 11.84VILKK156 pKa = 10.36LGSNVLKK163 pKa = 10.69QIGIGKK169 pKa = 9.8EE170 pKa = 3.94KK171 pKa = 10.8VQEE174 pKa = 4.06QNDD177 pKa = 3.36ALTRR181 pKa = 11.84FKK183 pKa = 10.06KK184 pKa = 9.23TKK186 pKa = 7.27TKK188 pKa = 9.9KK189 pKa = 9.06GRR191 pKa = 11.84VMEE194 pKa = 4.2VQFIEE199 pKa = 4.31SLKK202 pKa = 10.69GLEE205 pKa = 4.33AEE207 pKa = 4.51IGVDD211 pKa = 3.3KK212 pKa = 11.12NRR214 pKa = 11.84EE215 pKa = 3.36VDD217 pKa = 3.55TRR219 pKa = 11.84VSQSGVVLVTNDD231 pKa = 2.95RR232 pKa = 11.84RR233 pKa = 11.84YY234 pKa = 9.89VSQAHH239 pKa = 6.47VIFTCPTGDD248 pKa = 3.8TTWKK252 pKa = 7.91EE253 pKa = 4.07TARR256 pKa = 11.84QATKK260 pKa = 10.46RR261 pKa = 11.84SDD263 pKa = 2.65IRR265 pKa = 11.84AYY267 pKa = 10.8VHH269 pKa = 7.26DD270 pKa = 4.66PANGDD275 pKa = 3.72SEE277 pKa = 4.76KK278 pKa = 11.24GLLALVDD285 pKa = 4.25LLL287 pKa = 4.74
MM1 pKa = 7.37TRR3 pKa = 11.84LTLLAPGDD11 pKa = 4.07VIRR14 pKa = 11.84KK15 pKa = 9.61AEE17 pKa = 3.98QEE19 pKa = 4.01LKK21 pKa = 10.7ARR23 pKa = 11.84RR24 pKa = 11.84IDD26 pKa = 3.87VLIQNDD32 pKa = 3.65AEE34 pKa = 4.34EE35 pKa = 4.73GGKK38 pKa = 8.31EE39 pKa = 3.97QEE41 pKa = 4.13RR42 pKa = 11.84KK43 pKa = 10.36EE44 pKa = 3.82EE45 pKa = 4.24DD46 pKa = 3.16EE47 pKa = 4.75AGGKK51 pKa = 9.09RR52 pKa = 11.84VQEE55 pKa = 4.14STNEE59 pKa = 3.82RR60 pKa = 11.84KK61 pKa = 8.13EE62 pKa = 4.16TGNATEE68 pKa = 4.66RR69 pKa = 11.84GGEE72 pKa = 4.08GTQKK76 pKa = 10.62NDD78 pKa = 2.86EE79 pKa = 4.91DD80 pKa = 3.78GGEE83 pKa = 4.1RR84 pKa = 11.84QGRR87 pKa = 11.84EE88 pKa = 3.83AEE90 pKa = 3.92EE91 pKa = 4.1VQIHH95 pKa = 6.16KK96 pKa = 10.7AFSEE100 pKa = 4.19NKK102 pKa = 9.85GAGDD106 pKa = 4.32RR107 pKa = 11.84DD108 pKa = 4.24DD109 pKa = 5.04EE110 pKa = 4.51VQATKK115 pKa = 9.69QEE117 pKa = 4.12KK118 pKa = 9.94EE119 pKa = 4.12DD120 pKa = 3.75MVVLDD125 pKa = 4.61SEE127 pKa = 4.57VQKK130 pKa = 11.37VIDD133 pKa = 3.63KK134 pKa = 10.9LYY136 pKa = 10.69GIRR139 pKa = 11.84LQTIDD144 pKa = 4.41TITEE148 pKa = 3.54IGDD151 pKa = 3.68RR152 pKa = 11.84VILKK156 pKa = 10.36LGSNVLKK163 pKa = 10.69QIGIGKK169 pKa = 9.8EE170 pKa = 3.94KK171 pKa = 10.8VQEE174 pKa = 4.06QNDD177 pKa = 3.36ALTRR181 pKa = 11.84FKK183 pKa = 10.06KK184 pKa = 9.23TKK186 pKa = 7.27TKK188 pKa = 9.9KK189 pKa = 9.06GRR191 pKa = 11.84VMEE194 pKa = 4.2VQFIEE199 pKa = 4.31SLKK202 pKa = 10.69GLEE205 pKa = 4.33AEE207 pKa = 4.51IGVDD211 pKa = 3.3KK212 pKa = 11.12NRR214 pKa = 11.84EE215 pKa = 3.36VDD217 pKa = 3.55TRR219 pKa = 11.84VSQSGVVLVTNDD231 pKa = 2.95RR232 pKa = 11.84RR233 pKa = 11.84YY234 pKa = 9.89VSQAHH239 pKa = 6.47VIFTCPTGDD248 pKa = 3.8TTWKK252 pKa = 7.91EE253 pKa = 4.07TARR256 pKa = 11.84QATKK260 pKa = 10.46RR261 pKa = 11.84SDD263 pKa = 2.65IRR265 pKa = 11.84AYY267 pKa = 10.8VHH269 pKa = 7.26DD270 pKa = 4.66PANGDD275 pKa = 3.72SEE277 pKa = 4.76KK278 pKa = 11.24GLLALVDD285 pKa = 4.25LLL287 pKa = 4.74
Molecular weight: 32.07 kDa
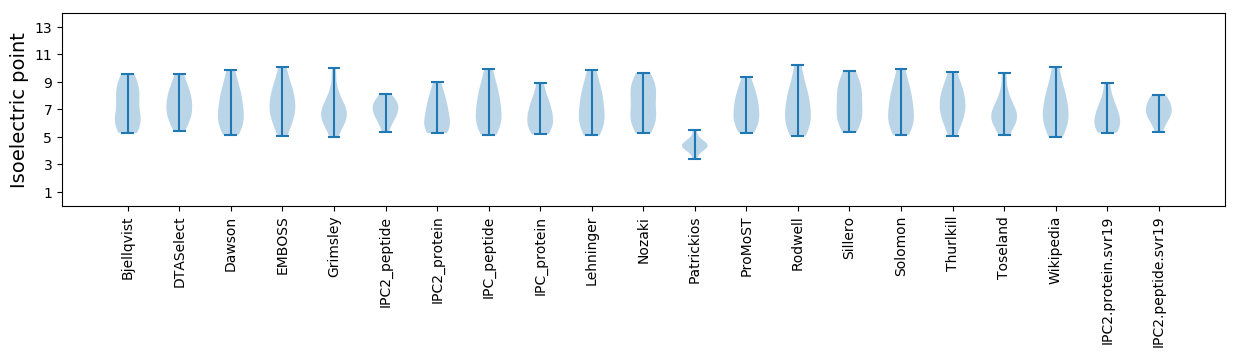
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M1H204|M1H204_9REOV Non-structural protein NS3 OS=Sathuvachari virus OX=1289939 PE=3 SV=1
MM1 pKa = 7.26MLTAVSHH8 pKa = 5.47IHH10 pKa = 6.14SGGSRR15 pKa = 11.84EE16 pKa = 4.18TRR18 pKa = 11.84DD19 pKa = 3.12MTAPMLPVSDD29 pKa = 5.25KK30 pKa = 9.69EE31 pKa = 4.26TSLNILTNALTSGTGSTAQQKK52 pKa = 9.24NEE54 pKa = 3.9KK55 pKa = 9.87ASYY58 pKa = 9.85GAIAEE63 pKa = 4.26ALRR66 pKa = 11.84DD67 pKa = 3.93EE68 pKa = 4.58PAVRR72 pKa = 11.84YY73 pKa = 9.34AKK75 pKa = 10.93YY76 pKa = 9.57EE77 pKa = 4.09VNSRR81 pKa = 11.84SIPKK85 pKa = 9.52VEE87 pKa = 3.94SRR89 pKa = 11.84IKK91 pKa = 10.39RR92 pKa = 11.84LRR94 pKa = 11.84RR95 pKa = 11.84TKK97 pKa = 10.26KK98 pKa = 9.13LYY100 pKa = 10.66KK101 pKa = 9.82IFEE104 pKa = 4.52VVFGVATLLLGILTTSMAEE123 pKa = 3.61IEE125 pKa = 4.19EE126 pKa = 4.48WKK128 pKa = 10.68DD129 pKa = 3.27KK130 pKa = 10.28PFKK133 pKa = 10.85GFASGLHH140 pKa = 5.0VVNLLMGAILLYY152 pKa = 9.99CAKK155 pKa = 10.2GVGKK159 pKa = 9.95IRR161 pKa = 11.84EE162 pKa = 4.1EE163 pKa = 3.76MRR165 pKa = 11.84QLEE168 pKa = 4.14RR169 pKa = 11.84DD170 pKa = 3.55LEE172 pKa = 4.23KK173 pKa = 10.85KK174 pKa = 10.23IMYY177 pKa = 10.03NSVAGRR183 pKa = 11.84TLGIQRR189 pKa = 11.84QHH191 pKa = 3.83VTMYY195 pKa = 10.37EE196 pKa = 4.03PANEE200 pKa = 4.31TMKK203 pKa = 10.63TMPLYY208 pKa = 6.74PTHH211 pKa = 6.27YY212 pKa = 10.27VV213 pKa = 3.09
MM1 pKa = 7.26MLTAVSHH8 pKa = 5.47IHH10 pKa = 6.14SGGSRR15 pKa = 11.84EE16 pKa = 4.18TRR18 pKa = 11.84DD19 pKa = 3.12MTAPMLPVSDD29 pKa = 5.25KK30 pKa = 9.69EE31 pKa = 4.26TSLNILTNALTSGTGSTAQQKK52 pKa = 9.24NEE54 pKa = 3.9KK55 pKa = 9.87ASYY58 pKa = 9.85GAIAEE63 pKa = 4.26ALRR66 pKa = 11.84DD67 pKa = 3.93EE68 pKa = 4.58PAVRR72 pKa = 11.84YY73 pKa = 9.34AKK75 pKa = 10.93YY76 pKa = 9.57EE77 pKa = 4.09VNSRR81 pKa = 11.84SIPKK85 pKa = 9.52VEE87 pKa = 3.94SRR89 pKa = 11.84IKK91 pKa = 10.39RR92 pKa = 11.84LRR94 pKa = 11.84RR95 pKa = 11.84TKK97 pKa = 10.26KK98 pKa = 9.13LYY100 pKa = 10.66KK101 pKa = 9.82IFEE104 pKa = 4.52VVFGVATLLLGILTTSMAEE123 pKa = 3.61IEE125 pKa = 4.19EE126 pKa = 4.48WKK128 pKa = 10.68DD129 pKa = 3.27KK130 pKa = 10.28PFKK133 pKa = 10.85GFASGLHH140 pKa = 5.0VVNLLMGAILLYY152 pKa = 9.99CAKK155 pKa = 10.2GVGKK159 pKa = 9.95IRR161 pKa = 11.84EE162 pKa = 4.1EE163 pKa = 3.76MRR165 pKa = 11.84QLEE168 pKa = 4.14RR169 pKa = 11.84DD170 pKa = 3.55LEE172 pKa = 4.23KK173 pKa = 10.85KK174 pKa = 10.23IMYY177 pKa = 10.03NSVAGRR183 pKa = 11.84TLGIQRR189 pKa = 11.84QHH191 pKa = 3.83VTMYY195 pKa = 10.37EE196 pKa = 4.03PANEE200 pKa = 4.31TMKK203 pKa = 10.63TMPLYY208 pKa = 6.74PTHH211 pKa = 6.27YY212 pKa = 10.27VV213 pKa = 3.09
Molecular weight: 23.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5962 |
213 |
1321 |
596.2 |
68.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.24 ± 0.511 | 1.342 ± 0.28 |
5.585 ± 0.407 | 7.296 ± 0.566 |
3.807 ± 0.217 | 5.938 ± 0.288 |
2.499 ± 0.211 | 6.189 ± 0.334 |
5.669 ± 0.638 | 9.074 ± 0.405 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.69 ± 0.328 | 4.143 ± 0.319 |
3.556 ± 0.419 | 3.975 ± 0.286 |
7.112 ± 0.406 | 6.156 ± 0.333 |
5.552 ± 0.242 | 7.045 ± 0.46 |
1.124 ± 0.276 | 3.992 ± 0.259 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
