
Compostimonas suwonensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Compostimonas
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
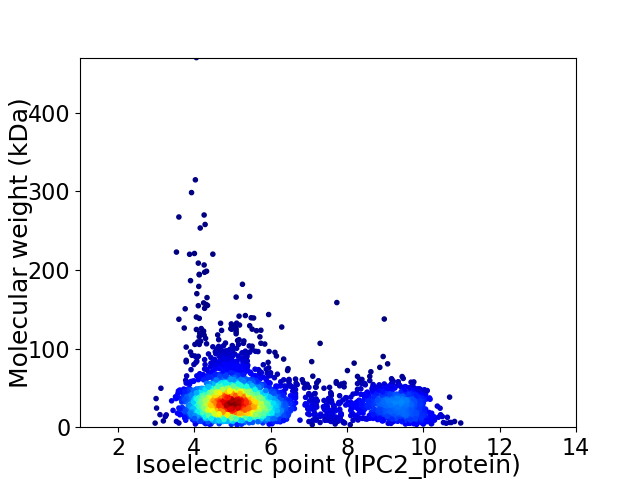
Virtual 2D-PAGE plot for 3422 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2M9C599|A0A2M9C599_9MICO Uncharacterized protein OS=Compostimonas suwonensis OX=1048394 GN=CLV54_0748 PE=4 SV=1
MM1 pKa = 7.58PRR3 pKa = 11.84LLFAGVLTAALALSPLAAQAANPQPTDD30 pKa = 3.66PASSTSTPATEE41 pKa = 4.3STATEE46 pKa = 4.59TPTSTAVPTEE56 pKa = 3.97ASTADD61 pKa = 3.6PTPAPGEE68 pKa = 4.17TPAPSEE74 pKa = 4.12TPAPGEE80 pKa = 4.17LPAPPPPPQPTTGAPAPGEE99 pKa = 4.02QTGTGTPTEE108 pKa = 4.35QNGLTAPAPAPGSAQRR124 pKa = 11.84AAVGPHH130 pKa = 6.45SISGTVTRR138 pKa = 11.84AGGAPVTAADD148 pKa = 4.22SVWVSLIDD156 pKa = 3.53QSSSSLIASTTTAIDD171 pKa = 3.71GSYY174 pKa = 9.2TLSDD178 pKa = 3.76IPSGDD183 pKa = 3.64DD184 pKa = 3.0MVVQFGSGSGTLLGEE199 pKa = 4.57YY200 pKa = 10.53YY201 pKa = 10.76DD202 pKa = 4.92DD203 pKa = 4.45APSAFFAQAFTFTDD217 pKa = 4.01EE218 pKa = 4.36EE219 pKa = 4.55PQTLTGIDD227 pKa = 3.5AEE229 pKa = 4.36LALGSVISGHH239 pKa = 5.7VSGSDD244 pKa = 3.25GSSVDD249 pKa = 3.31GVYY252 pKa = 11.19VNIQRR257 pKa = 11.84VPSGSGNSFFGGQVEE272 pKa = 4.09ADD274 pKa = 3.31GDD276 pKa = 4.03YY277 pKa = 10.86SITGVAPGDD286 pKa = 3.63YY287 pKa = 9.86RR288 pKa = 11.84VTFSGTPGGLMGQYY302 pKa = 10.58YY303 pKa = 10.8DD304 pKa = 3.74GVYY307 pKa = 10.54DD308 pKa = 3.54QGSATLVTVAGNGQTVTGIDD328 pKa = 4.15AILDD332 pKa = 4.02PGSSVSGTVTRR343 pKa = 11.84NDD345 pKa = 3.25GGSFEE350 pKa = 4.41TSPVTVSVQPVDD362 pKa = 3.59NQGFSVDD369 pKa = 3.32ATVAADD375 pKa = 3.94GSYY378 pKa = 9.38TVDD381 pKa = 3.03GLAPGVYY388 pKa = 8.44TVQFRR393 pKa = 11.84PQEE396 pKa = 4.17EE397 pKa = 4.68GSNLILGYY405 pKa = 10.71YY406 pKa = 10.22GGTDD410 pKa = 2.73WSTAARR416 pKa = 11.84ITIGAGEE423 pKa = 4.13AQDD426 pKa = 4.11LVGYY430 pKa = 10.15DD431 pKa = 3.57FGMLLGNEE439 pKa = 3.78ISGQITRR446 pKa = 11.84ADD448 pKa = 3.63GEE450 pKa = 4.04PWEE453 pKa = 4.44EE454 pKa = 3.75FRR456 pKa = 11.84TPQVIVEE463 pKa = 4.17EE464 pKa = 4.44WDD466 pKa = 3.71GSSTSVTSTSMEE478 pKa = 3.94ADD480 pKa = 2.94GSYY483 pKa = 10.33AVHH486 pKa = 6.98GIAPGSYY493 pKa = 7.62RR494 pKa = 11.84VKK496 pKa = 11.02FEE498 pKa = 4.05GSYY501 pKa = 11.15SDD503 pKa = 4.23DD504 pKa = 3.1AFGFVWYY511 pKa = 9.88DD512 pKa = 3.54DD513 pKa = 4.81VIDD516 pKa = 4.75PSAAHH521 pKa = 5.39QFVFGSDD528 pKa = 3.56PQVVTGIDD536 pKa = 3.63GQLSQASSVSGTVTRR551 pKa = 11.84SDD553 pKa = 3.15GKK555 pKa = 9.91PIEE558 pKa = 4.44EE559 pKa = 4.05NTFVSLEE566 pKa = 4.22LPGSGYY572 pKa = 10.04IAASTQVRR580 pKa = 11.84ADD582 pKa = 3.05GSYY585 pKa = 10.61RR586 pKa = 11.84FDD588 pKa = 3.16KK589 pKa = 9.61VTPRR593 pKa = 11.84DD594 pKa = 3.46YY595 pKa = 11.13VVRR598 pKa = 11.84FGGTDD603 pKa = 3.14HH604 pKa = 7.07VRR606 pKa = 11.84EE607 pKa = 3.92QYY609 pKa = 11.05YY610 pKa = 10.41PDD612 pKa = 3.27VQYY615 pKa = 11.16SYY617 pKa = 10.59EE618 pKa = 4.06ATILTVTEE626 pKa = 4.39EE627 pKa = 4.37PLTGIDD633 pKa = 3.32ATLTAGAPVTGTVTRR648 pKa = 11.84SDD650 pKa = 4.03GAPIDD655 pKa = 3.82YY656 pKa = 9.76TSIAVYY662 pKa = 9.96AYY664 pKa = 10.46DD665 pKa = 3.86ADD667 pKa = 3.82YY668 pKa = 10.97RR669 pKa = 11.84QAGSTWVNTDD679 pKa = 2.63GSYY682 pKa = 10.7KK683 pKa = 10.16IPALAAGNYY692 pKa = 8.86RR693 pKa = 11.84FQFTPLGTGNPVVPSWWDD711 pKa = 3.29GKK713 pKa = 11.19ASFEE717 pKa = 4.11QADD720 pKa = 4.32AVTMPSTLEE729 pKa = 3.88AVTGIDD735 pKa = 3.16TEE737 pKa = 5.15LIVGTTASGHH747 pKa = 4.96VVDD750 pKa = 5.41PNGDD754 pKa = 3.51PVANTSVSFAGADD767 pKa = 3.26GRR769 pKa = 11.84YY770 pKa = 10.46GYY772 pKa = 8.33GTTNADD778 pKa = 2.43GDD780 pKa = 4.2YY781 pKa = 10.24TVYY784 pKa = 11.18GLADD788 pKa = 3.54GDD790 pKa = 4.13YY791 pKa = 9.62TVQFSTWDD799 pKa = 2.98TGLIGEE805 pKa = 4.87YY806 pKa = 10.26YY807 pKa = 10.78DD808 pKa = 4.8DD809 pKa = 5.13ARR811 pKa = 11.84TVDD814 pKa = 4.09DD815 pKa = 3.85ATLVTVEE822 pKa = 4.01NAEE825 pKa = 4.16PVTGIDD831 pKa = 3.24AALEE835 pKa = 3.99QGSTISGTVTRR846 pKa = 11.84ADD848 pKa = 3.52GGSLADD854 pKa = 3.56CAVRR858 pKa = 11.84VSLVRR863 pKa = 11.84DD864 pKa = 3.89APPSQSGDD872 pKa = 3.03GGEE875 pKa = 4.84CVDD878 pKa = 4.57ADD880 pKa = 3.5GGYY883 pKa = 10.52AIDD886 pKa = 5.93GIAPDD891 pKa = 3.73RR892 pKa = 11.84YY893 pKa = 8.59FVAFGGEE900 pKa = 4.12GLATQWFDD908 pKa = 3.69DD909 pKa = 3.72VSVRR913 pKa = 11.84ADD915 pKa = 3.29ATVLEE920 pKa = 4.79FVEE923 pKa = 4.87GEE925 pKa = 4.03PRR927 pKa = 11.84DD928 pKa = 3.87EE929 pKa = 4.37TGVDD933 pKa = 3.56AEE935 pKa = 4.47LQPGGSITASATRR948 pKa = 11.84SDD950 pKa = 4.16GGSLDD955 pKa = 3.52AVTVEE960 pKa = 4.91AVLADD965 pKa = 3.89GGTVVYY971 pKa = 10.45SRR973 pKa = 11.84AVGSYY978 pKa = 9.27EE979 pKa = 3.85PQFTLSGLPLDD990 pKa = 4.54RR991 pKa = 11.84YY992 pKa = 10.19LLRR995 pKa = 11.84FSAPGLPTQYY1005 pKa = 10.9YY1006 pKa = 9.08GGADD1010 pKa = 3.19AGSATVVDD1018 pKa = 4.99LTTEE1022 pKa = 4.07PDD1024 pKa = 3.11ATGIAVALAAASTAALEE1041 pKa = 4.34VTRR1044 pKa = 11.84GGGEE1048 pKa = 4.2TVTAGQTLAALAVTVVDD1065 pKa = 4.3GAGEE1069 pKa = 3.88PVYY1072 pKa = 10.69GKK1074 pKa = 10.02RR1075 pKa = 11.84VVFTVSGAATFAGGATTATAPSNSAGLAVSPALLSGEE1112 pKa = 4.26TAGTVTASARR1122 pKa = 11.84ISGVSGSSVAFDD1134 pKa = 3.8PSTVRR1139 pKa = 11.84DD1140 pKa = 3.93AQDD1143 pKa = 3.19GAGDD1147 pKa = 4.14LSVTTTATTSLEE1159 pKa = 3.86NGLAVLTVTVLNSGPAAADD1178 pKa = 3.51LRR1180 pKa = 11.84ITTPFGAKK1188 pKa = 9.19TYY1190 pKa = 10.31TNVAPGAEE1198 pKa = 4.05ATRR1201 pKa = 11.84VFRR1204 pKa = 11.84SYY1206 pKa = 10.57RR1207 pKa = 11.84ASVDD1211 pKa = 3.43DD1212 pKa = 3.68GTVTVRR1218 pKa = 11.84VTDD1221 pKa = 3.73PVGGHH1226 pKa = 6.48SIDD1229 pKa = 3.59ISTDD1233 pKa = 3.41YY1234 pKa = 11.39EE1235 pKa = 4.52GVEE1238 pKa = 4.15LL1239 pKa = 5.16
MM1 pKa = 7.58PRR3 pKa = 11.84LLFAGVLTAALALSPLAAQAANPQPTDD30 pKa = 3.66PASSTSTPATEE41 pKa = 4.3STATEE46 pKa = 4.59TPTSTAVPTEE56 pKa = 3.97ASTADD61 pKa = 3.6PTPAPGEE68 pKa = 4.17TPAPSEE74 pKa = 4.12TPAPGEE80 pKa = 4.17LPAPPPPPQPTTGAPAPGEE99 pKa = 4.02QTGTGTPTEE108 pKa = 4.35QNGLTAPAPAPGSAQRR124 pKa = 11.84AAVGPHH130 pKa = 6.45SISGTVTRR138 pKa = 11.84AGGAPVTAADD148 pKa = 4.22SVWVSLIDD156 pKa = 3.53QSSSSLIASTTTAIDD171 pKa = 3.71GSYY174 pKa = 9.2TLSDD178 pKa = 3.76IPSGDD183 pKa = 3.64DD184 pKa = 3.0MVVQFGSGSGTLLGEE199 pKa = 4.57YY200 pKa = 10.53YY201 pKa = 10.76DD202 pKa = 4.92DD203 pKa = 4.45APSAFFAQAFTFTDD217 pKa = 4.01EE218 pKa = 4.36EE219 pKa = 4.55PQTLTGIDD227 pKa = 3.5AEE229 pKa = 4.36LALGSVISGHH239 pKa = 5.7VSGSDD244 pKa = 3.25GSSVDD249 pKa = 3.31GVYY252 pKa = 11.19VNIQRR257 pKa = 11.84VPSGSGNSFFGGQVEE272 pKa = 4.09ADD274 pKa = 3.31GDD276 pKa = 4.03YY277 pKa = 10.86SITGVAPGDD286 pKa = 3.63YY287 pKa = 9.86RR288 pKa = 11.84VTFSGTPGGLMGQYY302 pKa = 10.58YY303 pKa = 10.8DD304 pKa = 3.74GVYY307 pKa = 10.54DD308 pKa = 3.54QGSATLVTVAGNGQTVTGIDD328 pKa = 4.15AILDD332 pKa = 4.02PGSSVSGTVTRR343 pKa = 11.84NDD345 pKa = 3.25GGSFEE350 pKa = 4.41TSPVTVSVQPVDD362 pKa = 3.59NQGFSVDD369 pKa = 3.32ATVAADD375 pKa = 3.94GSYY378 pKa = 9.38TVDD381 pKa = 3.03GLAPGVYY388 pKa = 8.44TVQFRR393 pKa = 11.84PQEE396 pKa = 4.17EE397 pKa = 4.68GSNLILGYY405 pKa = 10.71YY406 pKa = 10.22GGTDD410 pKa = 2.73WSTAARR416 pKa = 11.84ITIGAGEE423 pKa = 4.13AQDD426 pKa = 4.11LVGYY430 pKa = 10.15DD431 pKa = 3.57FGMLLGNEE439 pKa = 3.78ISGQITRR446 pKa = 11.84ADD448 pKa = 3.63GEE450 pKa = 4.04PWEE453 pKa = 4.44EE454 pKa = 3.75FRR456 pKa = 11.84TPQVIVEE463 pKa = 4.17EE464 pKa = 4.44WDD466 pKa = 3.71GSSTSVTSTSMEE478 pKa = 3.94ADD480 pKa = 2.94GSYY483 pKa = 10.33AVHH486 pKa = 6.98GIAPGSYY493 pKa = 7.62RR494 pKa = 11.84VKK496 pKa = 11.02FEE498 pKa = 4.05GSYY501 pKa = 11.15SDD503 pKa = 4.23DD504 pKa = 3.1AFGFVWYY511 pKa = 9.88DD512 pKa = 3.54DD513 pKa = 4.81VIDD516 pKa = 4.75PSAAHH521 pKa = 5.39QFVFGSDD528 pKa = 3.56PQVVTGIDD536 pKa = 3.63GQLSQASSVSGTVTRR551 pKa = 11.84SDD553 pKa = 3.15GKK555 pKa = 9.91PIEE558 pKa = 4.44EE559 pKa = 4.05NTFVSLEE566 pKa = 4.22LPGSGYY572 pKa = 10.04IAASTQVRR580 pKa = 11.84ADD582 pKa = 3.05GSYY585 pKa = 10.61RR586 pKa = 11.84FDD588 pKa = 3.16KK589 pKa = 9.61VTPRR593 pKa = 11.84DD594 pKa = 3.46YY595 pKa = 11.13VVRR598 pKa = 11.84FGGTDD603 pKa = 3.14HH604 pKa = 7.07VRR606 pKa = 11.84EE607 pKa = 3.92QYY609 pKa = 11.05YY610 pKa = 10.41PDD612 pKa = 3.27VQYY615 pKa = 11.16SYY617 pKa = 10.59EE618 pKa = 4.06ATILTVTEE626 pKa = 4.39EE627 pKa = 4.37PLTGIDD633 pKa = 3.32ATLTAGAPVTGTVTRR648 pKa = 11.84SDD650 pKa = 4.03GAPIDD655 pKa = 3.82YY656 pKa = 9.76TSIAVYY662 pKa = 9.96AYY664 pKa = 10.46DD665 pKa = 3.86ADD667 pKa = 3.82YY668 pKa = 10.97RR669 pKa = 11.84QAGSTWVNTDD679 pKa = 2.63GSYY682 pKa = 10.7KK683 pKa = 10.16IPALAAGNYY692 pKa = 8.86RR693 pKa = 11.84FQFTPLGTGNPVVPSWWDD711 pKa = 3.29GKK713 pKa = 11.19ASFEE717 pKa = 4.11QADD720 pKa = 4.32AVTMPSTLEE729 pKa = 3.88AVTGIDD735 pKa = 3.16TEE737 pKa = 5.15LIVGTTASGHH747 pKa = 4.96VVDD750 pKa = 5.41PNGDD754 pKa = 3.51PVANTSVSFAGADD767 pKa = 3.26GRR769 pKa = 11.84YY770 pKa = 10.46GYY772 pKa = 8.33GTTNADD778 pKa = 2.43GDD780 pKa = 4.2YY781 pKa = 10.24TVYY784 pKa = 11.18GLADD788 pKa = 3.54GDD790 pKa = 4.13YY791 pKa = 9.62TVQFSTWDD799 pKa = 2.98TGLIGEE805 pKa = 4.87YY806 pKa = 10.26YY807 pKa = 10.78DD808 pKa = 4.8DD809 pKa = 5.13ARR811 pKa = 11.84TVDD814 pKa = 4.09DD815 pKa = 3.85ATLVTVEE822 pKa = 4.01NAEE825 pKa = 4.16PVTGIDD831 pKa = 3.24AALEE835 pKa = 3.99QGSTISGTVTRR846 pKa = 11.84ADD848 pKa = 3.52GGSLADD854 pKa = 3.56CAVRR858 pKa = 11.84VSLVRR863 pKa = 11.84DD864 pKa = 3.89APPSQSGDD872 pKa = 3.03GGEE875 pKa = 4.84CVDD878 pKa = 4.57ADD880 pKa = 3.5GGYY883 pKa = 10.52AIDD886 pKa = 5.93GIAPDD891 pKa = 3.73RR892 pKa = 11.84YY893 pKa = 8.59FVAFGGEE900 pKa = 4.12GLATQWFDD908 pKa = 3.69DD909 pKa = 3.72VSVRR913 pKa = 11.84ADD915 pKa = 3.29ATVLEE920 pKa = 4.79FVEE923 pKa = 4.87GEE925 pKa = 4.03PRR927 pKa = 11.84DD928 pKa = 3.87EE929 pKa = 4.37TGVDD933 pKa = 3.56AEE935 pKa = 4.47LQPGGSITASATRR948 pKa = 11.84SDD950 pKa = 4.16GGSLDD955 pKa = 3.52AVTVEE960 pKa = 4.91AVLADD965 pKa = 3.89GGTVVYY971 pKa = 10.45SRR973 pKa = 11.84AVGSYY978 pKa = 9.27EE979 pKa = 3.85PQFTLSGLPLDD990 pKa = 4.54RR991 pKa = 11.84YY992 pKa = 10.19LLRR995 pKa = 11.84FSAPGLPTQYY1005 pKa = 10.9YY1006 pKa = 9.08GGADD1010 pKa = 3.19AGSATVVDD1018 pKa = 4.99LTTEE1022 pKa = 4.07PDD1024 pKa = 3.11ATGIAVALAAASTAALEE1041 pKa = 4.34VTRR1044 pKa = 11.84GGGEE1048 pKa = 4.2TVTAGQTLAALAVTVVDD1065 pKa = 4.3GAGEE1069 pKa = 3.88PVYY1072 pKa = 10.69GKK1074 pKa = 10.02RR1075 pKa = 11.84VVFTVSGAATFAGGATTATAPSNSAGLAVSPALLSGEE1112 pKa = 4.26TAGTVTASARR1122 pKa = 11.84ISGVSGSSVAFDD1134 pKa = 3.8PSTVRR1139 pKa = 11.84DD1140 pKa = 3.93AQDD1143 pKa = 3.19GAGDD1147 pKa = 4.14LSVTTTATTSLEE1159 pKa = 3.86NGLAVLTVTVLNSGPAAADD1178 pKa = 3.51LRR1180 pKa = 11.84ITTPFGAKK1188 pKa = 9.19TYY1190 pKa = 10.31TNVAPGAEE1198 pKa = 4.05ATRR1201 pKa = 11.84VFRR1204 pKa = 11.84SYY1206 pKa = 10.57RR1207 pKa = 11.84ASVDD1211 pKa = 3.43DD1212 pKa = 3.68GTVTVRR1218 pKa = 11.84VTDD1221 pKa = 3.73PVGGHH1226 pKa = 6.48SIDD1229 pKa = 3.59ISTDD1233 pKa = 3.41YY1234 pKa = 11.39EE1235 pKa = 4.52GVEE1238 pKa = 4.15LL1239 pKa = 5.16
Molecular weight: 126.09 kDa
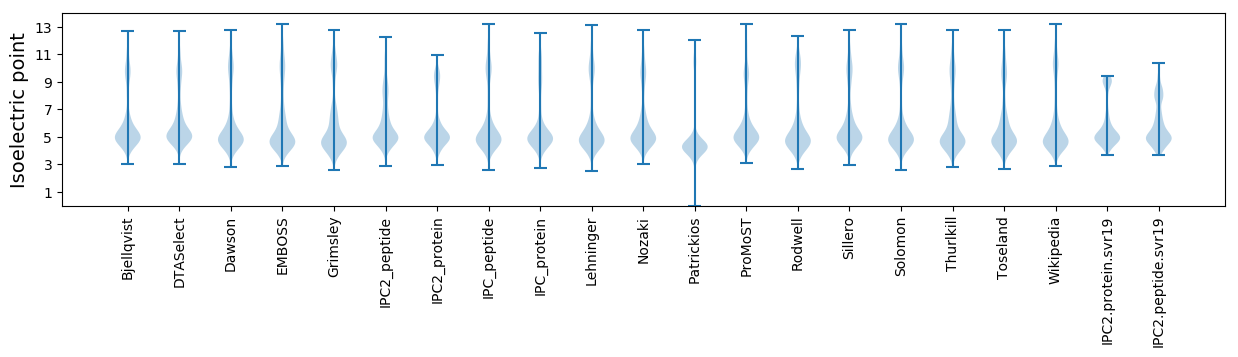
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2M9C4F9|A0A2M9C4F9_9MICO Signal transduction histidine kinase OS=Compostimonas suwonensis OX=1048394 GN=CLV54_0448 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.98GRR40 pKa = 11.84TEE42 pKa = 3.99LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.98GRR40 pKa = 11.84TEE42 pKa = 3.99LSAA45 pKa = 4.86
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1170212 |
29 |
4639 |
342.0 |
36.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.998 ± 0.056 | 0.465 ± 0.01 |
6.15 ± 0.043 | 5.657 ± 0.037 |
3.169 ± 0.023 | 9.129 ± 0.043 |
1.872 ± 0.023 | 4.619 ± 0.036 |
1.857 ± 0.026 | 10.06 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.656 ± 0.019 | 2.154 ± 0.028 |
5.523 ± 0.035 | 2.846 ± 0.023 |
6.84 ± 0.057 | 6.249 ± 0.034 |
6.313 ± 0.051 | 8.879 ± 0.038 |
1.473 ± 0.016 | 2.092 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
