
Ethiopian tobacco bushy top virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Calvusvirinae; Umbravirus
Average proteome isoelectric point is 7.36
Get precalculated fractions of proteins
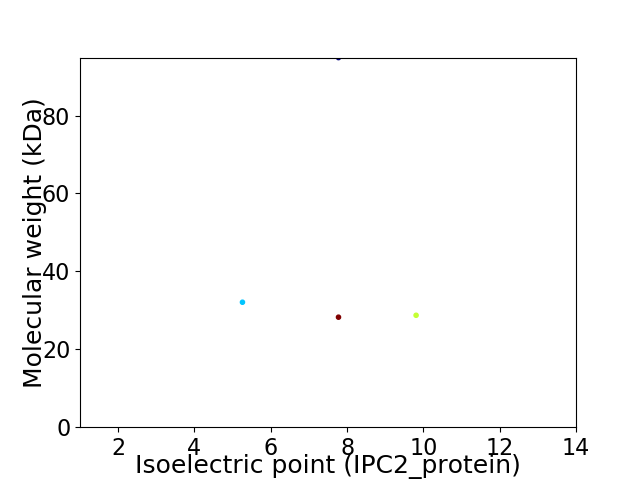
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L6UUR6|A0A1L6UUR6_9TOMB Replication-associated protein OS=Ethiopian tobacco bushy top virus OX=1538549 PE=4 SV=1
MM1 pKa = 8.1AIRR4 pKa = 11.84AITNWLRR11 pKa = 11.84LDD13 pKa = 3.83YY14 pKa = 11.33SPAPYY19 pKa = 9.36IKK21 pKa = 10.08SRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84CNEE28 pKa = 3.46DD29 pKa = 3.13YY30 pKa = 11.47GEE32 pKa = 3.97XWGLLITQARR42 pKa = 11.84MTEE45 pKa = 4.45SAGHH49 pKa = 6.69HH50 pKa = 4.69MMAWYY55 pKa = 9.59EE56 pKa = 4.04GSAEE60 pKa = 4.23VNGLIPGHH68 pKa = 7.24DD69 pKa = 4.14DD70 pKa = 4.03QGLADD75 pKa = 4.55PAPPPVMEE83 pKa = 4.3VTTPPNEE90 pKa = 4.16EE91 pKa = 4.12VSCPGPSDD99 pKa = 3.4DD100 pKa = 6.1HH101 pKa = 7.49GVEE104 pKa = 3.97WVEE107 pKa = 4.23NLPSPPSPKK116 pKa = 9.89DD117 pKa = 3.08QRR119 pKa = 11.84NRR121 pKa = 11.84LPTLXEE127 pKa = 3.95VLGADD132 pKa = 3.79RR133 pKa = 11.84LQIIPYY139 pKa = 8.39TGGHH143 pKa = 5.7RR144 pKa = 11.84VLSEE148 pKa = 3.84XEE150 pKa = 4.06VRR152 pKa = 11.84PRR154 pKa = 11.84AQGVILLPAPRR165 pKa = 11.84YY166 pKa = 9.65RR167 pKa = 11.84PSLLEE172 pKa = 3.88RR173 pKa = 11.84ALDD176 pKa = 3.6ALVFAWRR183 pKa = 11.84PASQALDD190 pKa = 3.44ALVRR194 pKa = 11.84SQPQEE199 pKa = 4.16DD200 pKa = 4.61GLPASLKK207 pKa = 9.89LQEE210 pKa = 4.32EE211 pKa = 4.91GAPXEE216 pKa = 4.85YY217 pKa = 8.93ITAHH221 pKa = 6.42MIAMEE226 pKa = 4.33LRR228 pKa = 11.84SMFGWQLNTPANRR241 pKa = 11.84EE242 pKa = 3.85LGNRR246 pKa = 11.84VARR249 pKa = 11.84DD250 pKa = 3.42ILRR253 pKa = 11.84DD254 pKa = 3.19RR255 pKa = 11.84CGANRR260 pKa = 11.84EE261 pKa = 4.22NTWYY265 pKa = 10.56LSSLGVQMWFQPTLCDD281 pKa = 3.52LAVKK285 pKa = 10.49AGPQNFF291 pKa = 3.45
MM1 pKa = 8.1AIRR4 pKa = 11.84AITNWLRR11 pKa = 11.84LDD13 pKa = 3.83YY14 pKa = 11.33SPAPYY19 pKa = 9.36IKK21 pKa = 10.08SRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84CNEE28 pKa = 3.46DD29 pKa = 3.13YY30 pKa = 11.47GEE32 pKa = 3.97XWGLLITQARR42 pKa = 11.84MTEE45 pKa = 4.45SAGHH49 pKa = 6.69HH50 pKa = 4.69MMAWYY55 pKa = 9.59EE56 pKa = 4.04GSAEE60 pKa = 4.23VNGLIPGHH68 pKa = 7.24DD69 pKa = 4.14DD70 pKa = 4.03QGLADD75 pKa = 4.55PAPPPVMEE83 pKa = 4.3VTTPPNEE90 pKa = 4.16EE91 pKa = 4.12VSCPGPSDD99 pKa = 3.4DD100 pKa = 6.1HH101 pKa = 7.49GVEE104 pKa = 3.97WVEE107 pKa = 4.23NLPSPPSPKK116 pKa = 9.89DD117 pKa = 3.08QRR119 pKa = 11.84NRR121 pKa = 11.84LPTLXEE127 pKa = 3.95VLGADD132 pKa = 3.79RR133 pKa = 11.84LQIIPYY139 pKa = 8.39TGGHH143 pKa = 5.7RR144 pKa = 11.84VLSEE148 pKa = 3.84XEE150 pKa = 4.06VRR152 pKa = 11.84PRR154 pKa = 11.84AQGVILLPAPRR165 pKa = 11.84YY166 pKa = 9.65RR167 pKa = 11.84PSLLEE172 pKa = 3.88RR173 pKa = 11.84ALDD176 pKa = 3.6ALVFAWRR183 pKa = 11.84PASQALDD190 pKa = 3.44ALVRR194 pKa = 11.84SQPQEE199 pKa = 4.16DD200 pKa = 4.61GLPASLKK207 pKa = 9.89LQEE210 pKa = 4.32EE211 pKa = 4.91GAPXEE216 pKa = 4.85YY217 pKa = 8.93ITAHH221 pKa = 6.42MIAMEE226 pKa = 4.33LRR228 pKa = 11.84SMFGWQLNTPANRR241 pKa = 11.84EE242 pKa = 3.85LGNRR246 pKa = 11.84VARR249 pKa = 11.84DD250 pKa = 3.42ILRR253 pKa = 11.84DD254 pKa = 3.19RR255 pKa = 11.84CGANRR260 pKa = 11.84EE261 pKa = 4.22NTWYY265 pKa = 10.56LSSLGVQMWFQPTLCDD281 pKa = 3.52LAVKK285 pKa = 10.49AGPQNFF291 pKa = 3.45
Molecular weight: 32.08 kDa
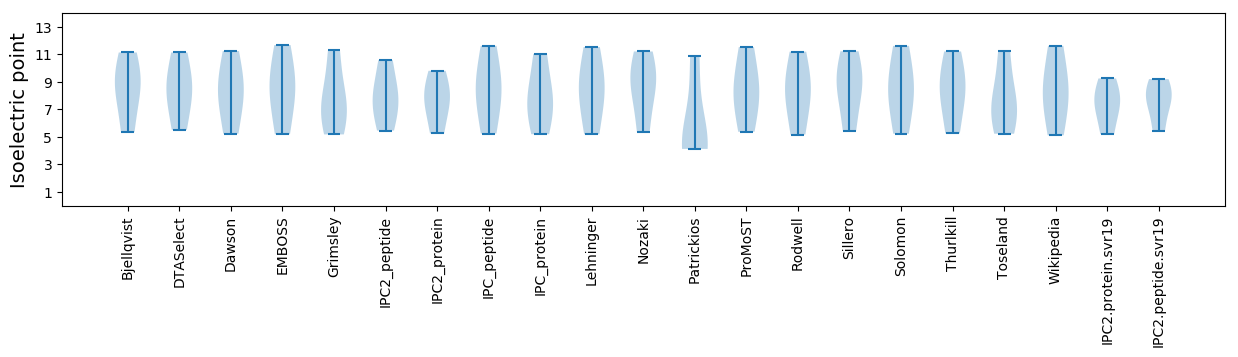
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L6UUR6|A0A1L6UUR6_9TOMB Replication-associated protein OS=Ethiopian tobacco bushy top virus OX=1538549 PE=4 SV=1
MM1 pKa = 7.83AEE3 pKa = 4.22HH4 pKa = 7.06SLLCLVTGRR13 pKa = 11.84PARR16 pKa = 11.84LAADD20 pKa = 3.43NGRR23 pKa = 11.84WVSMTCNINVYY34 pKa = 9.31TSNKK38 pKa = 9.13SSNSRR43 pKa = 11.84RR44 pKa = 11.84APRR47 pKa = 11.84GPVWRR52 pKa = 11.84GHH54 pKa = 5.18RR55 pKa = 11.84QGASRR60 pKa = 11.84NRR62 pKa = 11.84PGRR65 pKa = 11.84VNASQRR71 pKa = 11.84RR72 pKa = 11.84PEE74 pKa = 4.34GGVYY78 pKa = 9.96PPPAPKK84 pKa = 8.79NTKK87 pKa = 9.44QNSGGVAQVPPNPKK101 pKa = 9.44HH102 pKa = 6.55RR103 pKa = 11.84GATIHH108 pKa = 6.9RR109 pKa = 11.84EE110 pKa = 4.26SGCGIHH116 pKa = 6.82PPRR119 pKa = 11.84SRR121 pKa = 11.84RR122 pKa = 11.84CARR125 pKa = 11.84RR126 pKa = 11.84SRR128 pKa = 11.84SVDD131 pKa = 2.94PRR133 pKa = 11.84RR134 pKa = 11.84PTPKK138 pKa = 10.08PKK140 pKa = 10.09QRR142 pKa = 11.84WAAAEE147 pKa = 3.74ISAEE151 pKa = 3.76RR152 pKa = 11.84RR153 pKa = 11.84AEE155 pKa = 3.84IDD157 pKa = 3.5GLLSPLFDD165 pKa = 3.34TLGRR169 pKa = 11.84QVQGDD174 pKa = 3.92AAVLRR179 pKa = 11.84YY180 pKa = 10.09CFGATQRR187 pKa = 11.84QLRR190 pKa = 11.84KK191 pKa = 8.34WRR193 pKa = 11.84KK194 pKa = 7.92PVQPAYY200 pKa = 10.8NVAAPNRR207 pKa = 11.84EE208 pKa = 3.91CSAQLSSTATTNAEE222 pKa = 4.1DD223 pKa = 3.83LQGDD227 pKa = 3.98GKK229 pKa = 11.06GRR231 pKa = 11.84SGQLDD236 pKa = 3.51KK237 pKa = 11.85SEE239 pKa = 4.21VLHH242 pKa = 6.76SGSDD246 pKa = 3.63VPKK249 pKa = 10.21VHH251 pKa = 6.77TDD253 pKa = 3.39CGRR256 pKa = 11.84TATNKK261 pKa = 7.75WW262 pKa = 3.12
MM1 pKa = 7.83AEE3 pKa = 4.22HH4 pKa = 7.06SLLCLVTGRR13 pKa = 11.84PARR16 pKa = 11.84LAADD20 pKa = 3.43NGRR23 pKa = 11.84WVSMTCNINVYY34 pKa = 9.31TSNKK38 pKa = 9.13SSNSRR43 pKa = 11.84RR44 pKa = 11.84APRR47 pKa = 11.84GPVWRR52 pKa = 11.84GHH54 pKa = 5.18RR55 pKa = 11.84QGASRR60 pKa = 11.84NRR62 pKa = 11.84PGRR65 pKa = 11.84VNASQRR71 pKa = 11.84RR72 pKa = 11.84PEE74 pKa = 4.34GGVYY78 pKa = 9.96PPPAPKK84 pKa = 8.79NTKK87 pKa = 9.44QNSGGVAQVPPNPKK101 pKa = 9.44HH102 pKa = 6.55RR103 pKa = 11.84GATIHH108 pKa = 6.9RR109 pKa = 11.84EE110 pKa = 4.26SGCGIHH116 pKa = 6.82PPRR119 pKa = 11.84SRR121 pKa = 11.84RR122 pKa = 11.84CARR125 pKa = 11.84RR126 pKa = 11.84SRR128 pKa = 11.84SVDD131 pKa = 2.94PRR133 pKa = 11.84RR134 pKa = 11.84PTPKK138 pKa = 10.08PKK140 pKa = 10.09QRR142 pKa = 11.84WAAAEE147 pKa = 3.74ISAEE151 pKa = 3.76RR152 pKa = 11.84RR153 pKa = 11.84AEE155 pKa = 3.84IDD157 pKa = 3.5GLLSPLFDD165 pKa = 3.34TLGRR169 pKa = 11.84QVQGDD174 pKa = 3.92AAVLRR179 pKa = 11.84YY180 pKa = 10.09CFGATQRR187 pKa = 11.84QLRR190 pKa = 11.84KK191 pKa = 8.34WRR193 pKa = 11.84KK194 pKa = 7.92PVQPAYY200 pKa = 10.8NVAAPNRR207 pKa = 11.84EE208 pKa = 3.91CSAQLSSTATTNAEE222 pKa = 4.1DD223 pKa = 3.83LQGDD227 pKa = 3.98GKK229 pKa = 11.06GRR231 pKa = 11.84SGQLDD236 pKa = 3.51KK237 pKa = 11.85SEE239 pKa = 4.21VLHH242 pKa = 6.76SGSDD246 pKa = 3.63VPKK249 pKa = 10.21VHH251 pKa = 6.77TDD253 pKa = 3.39CGRR256 pKa = 11.84TATNKK261 pKa = 7.75WW262 pKa = 3.12
Molecular weight: 28.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1656 |
258 |
845 |
414.0 |
45.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.971 ± 0.661 | 2.114 ± 0.262 |
4.831 ± 0.288 | 5.556 ± 0.505 |
2.657 ± 0.604 | 7.367 ± 0.485 |
2.415 ± 0.095 | 3.744 ± 0.371 |
3.623 ± 0.614 | 8.877 ± 0.77 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.536 ± 0.335 | 4.408 ± 0.479 |
8.092 ± 0.729 | 3.925 ± 0.457 |
8.756 ± 0.827 | 6.401 ± 0.523 |
4.408 ± 0.681 | 6.944 ± 0.569 |
1.993 ± 0.347 | 2.778 ± 0.434 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
