
Hydrogenophaga sp. BA0156
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Hydrogenophaga; unclassified Hydrogenophaga
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
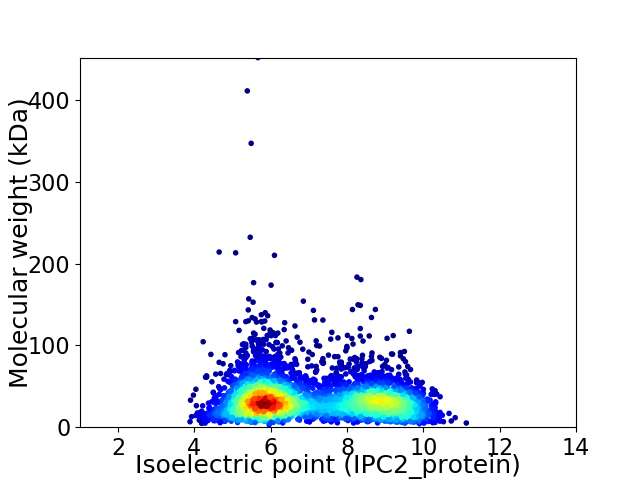
Virtual 2D-PAGE plot for 4261 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G8IIM5|A0A6G8IIM5_9BURK Uncharacterized protein OS=Hydrogenophaga sp. BA0156 OX=2716225 GN=G9Q37_13040 PE=4 SV=1
MM1 pKa = 7.48SSSPSEE7 pKa = 4.26PDD9 pKa = 3.84SPDD12 pKa = 3.32QPGEE16 pKa = 3.85ASLPCVMSFNANDD29 pKa = 3.73PSGAGGLSADD39 pKa = 3.97LTAMSSASAHH49 pKa = 5.36VLPVVTGTYY58 pKa = 10.45VRR60 pKa = 11.84DD61 pKa = 3.63TTAVRR66 pKa = 11.84DD67 pKa = 4.14HH68 pKa = 6.73IAFDD72 pKa = 3.76EE73 pKa = 4.29EE74 pKa = 4.4AVLDD78 pKa = 3.9QARR81 pKa = 11.84CALEE85 pKa = 4.41DD86 pKa = 3.59MPVQAFKK93 pKa = 11.21VGFVGTPEE101 pKa = 4.06NLSAIAQITSDD112 pKa = 3.77YY113 pKa = 11.41AEE115 pKa = 4.48VPVITYY121 pKa = 9.06MPDD124 pKa = 3.44LSWWDD129 pKa = 3.43EE130 pKa = 4.23LEE132 pKa = 4.17IEE134 pKa = 4.71TYY136 pKa = 11.2LDD138 pKa = 3.44ACTEE142 pKa = 4.14LLLSQTTVLVGNHH155 pKa = 5.0STLSRR160 pKa = 11.84WLLPDD165 pKa = 3.28WEE167 pKa = 4.82GDD169 pKa = 3.48RR170 pKa = 11.84PPGPRR175 pKa = 11.84EE176 pKa = 3.73VARR179 pKa = 11.84AAAVHH184 pKa = 5.67GVSYY188 pKa = 9.71TLVTGFNAADD198 pKa = 3.46QFLEE202 pKa = 4.04SHH204 pKa = 6.76LASPEE209 pKa = 4.24TVHH212 pKa = 6.13ATARR216 pKa = 11.84YY217 pKa = 8.64EE218 pKa = 3.96RR219 pKa = 11.84FEE221 pKa = 4.21ATFSGAGDD229 pKa = 3.74TLSAALCALIAGGADD244 pKa = 3.51LQNACAEE251 pKa = 4.13ALTYY255 pKa = 10.72LDD257 pKa = 3.41QCLDD261 pKa = 3.39AGFQPGMGNAVPDD274 pKa = 3.88RR275 pKa = 11.84LFWAHH280 pKa = 7.54DD281 pKa = 3.82DD282 pKa = 3.94DD283 pKa = 4.96EE284 pKa = 5.21EE285 pKa = 6.0PGGAPAAEE293 pKa = 4.28PASDD297 pKa = 3.71STLDD301 pKa = 4.15ADD303 pKa = 5.84DD304 pKa = 5.23FPLDD308 pKa = 3.9TTRR311 pKa = 11.84HH312 pKa = 4.6
MM1 pKa = 7.48SSSPSEE7 pKa = 4.26PDD9 pKa = 3.84SPDD12 pKa = 3.32QPGEE16 pKa = 3.85ASLPCVMSFNANDD29 pKa = 3.73PSGAGGLSADD39 pKa = 3.97LTAMSSASAHH49 pKa = 5.36VLPVVTGTYY58 pKa = 10.45VRR60 pKa = 11.84DD61 pKa = 3.63TTAVRR66 pKa = 11.84DD67 pKa = 4.14HH68 pKa = 6.73IAFDD72 pKa = 3.76EE73 pKa = 4.29EE74 pKa = 4.4AVLDD78 pKa = 3.9QARR81 pKa = 11.84CALEE85 pKa = 4.41DD86 pKa = 3.59MPVQAFKK93 pKa = 11.21VGFVGTPEE101 pKa = 4.06NLSAIAQITSDD112 pKa = 3.77YY113 pKa = 11.41AEE115 pKa = 4.48VPVITYY121 pKa = 9.06MPDD124 pKa = 3.44LSWWDD129 pKa = 3.43EE130 pKa = 4.23LEE132 pKa = 4.17IEE134 pKa = 4.71TYY136 pKa = 11.2LDD138 pKa = 3.44ACTEE142 pKa = 4.14LLLSQTTVLVGNHH155 pKa = 5.0STLSRR160 pKa = 11.84WLLPDD165 pKa = 3.28WEE167 pKa = 4.82GDD169 pKa = 3.48RR170 pKa = 11.84PPGPRR175 pKa = 11.84EE176 pKa = 3.73VARR179 pKa = 11.84AAAVHH184 pKa = 5.67GVSYY188 pKa = 9.71TLVTGFNAADD198 pKa = 3.46QFLEE202 pKa = 4.04SHH204 pKa = 6.76LASPEE209 pKa = 4.24TVHH212 pKa = 6.13ATARR216 pKa = 11.84YY217 pKa = 8.64EE218 pKa = 3.96RR219 pKa = 11.84FEE221 pKa = 4.21ATFSGAGDD229 pKa = 3.74TLSAALCALIAGGADD244 pKa = 3.51LQNACAEE251 pKa = 4.13ALTYY255 pKa = 10.72LDD257 pKa = 3.41QCLDD261 pKa = 3.39AGFQPGMGNAVPDD274 pKa = 3.88RR275 pKa = 11.84LFWAHH280 pKa = 7.54DD281 pKa = 3.82DD282 pKa = 3.94DD283 pKa = 4.96EE284 pKa = 5.21EE285 pKa = 6.0PGGAPAAEE293 pKa = 4.28PASDD297 pKa = 3.71STLDD301 pKa = 4.15ADD303 pKa = 5.84DD304 pKa = 5.23FPLDD308 pKa = 3.9TTRR311 pKa = 11.84HH312 pKa = 4.6
Molecular weight: 33.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G8ILB7|A0A6G8ILB7_9BURK 5-carboxymethyl-2-hydroxymuconate semialdehyde dehydrogenase OS=Hydrogenophaga sp. BA0156 OX=2716225 GN=hpaE PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.07IRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.91GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.07IRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.91GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1407824 |
28 |
4226 |
330.4 |
35.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.627 ± 0.065 | 0.892 ± 0.012 |
5.11 ± 0.025 | 5.248 ± 0.034 |
3.364 ± 0.022 | 8.47 ± 0.036 |
2.397 ± 0.02 | 3.865 ± 0.028 |
2.808 ± 0.029 | 11.223 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.291 ± 0.02 | 2.396 ± 0.021 |
5.591 ± 0.03 | 4.09 ± 0.026 |
7.551 ± 0.035 | 4.861 ± 0.026 |
4.936 ± 0.022 | 7.721 ± 0.03 |
1.581 ± 0.019 | 1.979 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
