
Phyllobacterium zundukense
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Phyllobacterium
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
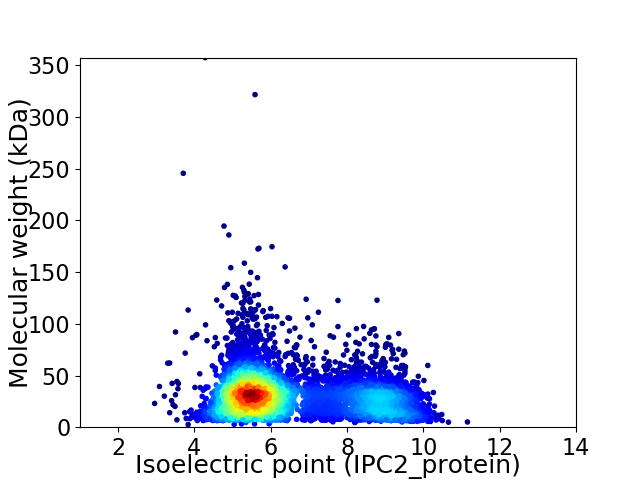
Virtual 2D-PAGE plot for 5605 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N9VW59|A0A2N9VW59_9RHIZ Uncharacterized protein OS=Phyllobacterium zundukense OX=1867719 GN=B5P45_17710 PE=4 SV=1
MM1 pKa = 7.39APAVAFADD9 pKa = 4.18CVTLGGVITCDD20 pKa = 3.03TAAPNPFTSTVGAGNTAAADD40 pKa = 3.98NITVNVGPGAQVAVGNLNAISLRR63 pKa = 11.84NNAVITIAGGGLVSATATSTTGNYY87 pKa = 7.37NTGGNAVEE95 pKa = 4.39VNNNGTLTIEE105 pKa = 4.4QGGQLLALGTQGSAEE120 pKa = 3.82AVNFQGTGNVLTNNGTIDD138 pKa = 3.65ANNSVAIWSQNTSGLNTVINNEE160 pKa = 4.0TGIIEE165 pKa = 4.59AGNGTTSTVIGGSGAGALDD184 pKa = 3.52FTNRR188 pKa = 11.84GTIRR192 pKa = 11.84GSINLANGNDD202 pKa = 3.4ILRR205 pKa = 11.84LYY207 pKa = 9.46TGSTVTGNFSGGGGTDD223 pKa = 4.04AIFLSGDD230 pKa = 3.77GQSSMAGNFNGFEE243 pKa = 4.18SLVKK247 pKa = 10.44NDD249 pKa = 3.7LGTWTLTGTITGVTVATVQNGTLVLTGANTNYY281 pKa = 9.01TGQVIIDD288 pKa = 3.6PAGTLEE294 pKa = 4.5ARR296 pKa = 11.84AQALPTQLLPANNVANITNNGLVRR320 pKa = 11.84FAQPDD325 pKa = 3.12NGTYY329 pKa = 10.03VGQIVGTGAVEE340 pKa = 3.87KK341 pKa = 9.83TGAGTLTLEE350 pKa = 4.39PSVAAGNTYY359 pKa = 10.78SGGTFINGGVVAVGADD375 pKa = 3.45NALGASTGGVSFNGGTLQLNSSFNLSAGRR404 pKa = 11.84AVTLNVGGGTINTQAFDD421 pKa = 3.38STLAQGVTGPGGLTKK436 pKa = 10.68AGAGTLTLTGNSDD449 pKa = 3.54YY450 pKa = 11.64SGGTVISAGTLQLGDD465 pKa = 4.02SGTSGSISGDD475 pKa = 3.33VLNNSLLAFNRR486 pKa = 11.84SDD488 pKa = 3.23AFLVPGVISGSGAVEE503 pKa = 3.92QNGSGTTILTGNNTYY518 pKa = 10.94SGGTTINAGTLQLGNGGTSGSIVGDD543 pKa = 3.6VANDD547 pKa = 3.39GSLAFNRR554 pKa = 11.84SDD556 pKa = 3.16STNFDD561 pKa = 3.59GVISSAGNVRR571 pKa = 11.84QIGGGTTILTAQNTYY586 pKa = 10.87SGATTVEE593 pKa = 4.03AGTLAAGATNVFSPNSSTSVLTGGTLDD620 pKa = 4.32AAGFDD625 pKa = 3.77QTVSALTNAGLVNVGGAPGTTLTVSGNYY653 pKa = 9.6VGADD657 pKa = 3.05GTLRR661 pKa = 11.84LNTALGDD668 pKa = 4.04DD669 pKa = 3.94ASTTDD674 pKa = 3.42RR675 pKa = 11.84LVVLGDD681 pKa = 3.67TSGSTTLVVANVGGSGAPTVEE702 pKa = 4.91GIRR705 pKa = 11.84VIDD708 pKa = 3.73VAGSSAGSFALDD720 pKa = 2.68GDD722 pKa = 4.33YY723 pKa = 11.36AFEE726 pKa = 4.8GDD728 pKa = 3.52EE729 pKa = 4.03AVIGGAYY736 pKa = 10.14AYY738 pKa = 10.24RR739 pKa = 11.84LYY741 pKa = 11.03KK742 pKa = 10.63NGIATPTDD750 pKa = 3.69GNWYY754 pKa = 9.8LRR756 pKa = 11.84SALVDD761 pKa = 3.64PADD764 pKa = 3.86PTAPTDD770 pKa = 3.67PTDD773 pKa = 3.87PTDD776 pKa = 3.89PTDD779 pKa = 3.76PTSPSSPLYY788 pKa = 9.64QPGVPVYY795 pKa = 9.58EE796 pKa = 5.31AYY798 pKa = 10.36AQTLLGLNGLPTLQQRR814 pKa = 11.84VGNRR818 pKa = 11.84YY819 pKa = 8.54WSGAGNGMLSQGDD832 pKa = 4.11GPGTIEE838 pKa = 4.97PAPQPSDD845 pKa = 3.12GGPAFVEE852 pKa = 5.1GRR854 pKa = 11.84GLWARR859 pKa = 11.84IEE861 pKa = 4.2GAHH864 pKa = 6.62GNFDD868 pKa = 3.87PRR870 pKa = 11.84TSTSSTDD877 pKa = 3.23YY878 pKa = 11.57DD879 pKa = 4.26LDD881 pKa = 3.3TWKK884 pKa = 10.27IQSGLDD890 pKa = 3.37GQLYY894 pKa = 9.89EE895 pKa = 4.52SEE897 pKa = 4.19SGKK900 pKa = 10.92LIGGITAQYY909 pKa = 10.11GNASADD915 pKa = 3.5VSSLFGNGEE924 pKa = 4.02VDD926 pKa = 2.95TDD928 pKa = 4.33GYY930 pKa = 11.6GVGATLTWYY939 pKa = 9.28GQNGFYY945 pKa = 10.78LDD947 pKa = 4.01GQAQATWYY955 pKa = 10.24EE956 pKa = 4.13SDD958 pKa = 3.81LSSGVLGSLSNGNDD972 pKa = 3.04GFGYY976 pKa = 10.62AFSLEE981 pKa = 4.2TGKK984 pKa = 10.72RR985 pKa = 11.84LALNQNWTLTPQAQLAYY1002 pKa = 10.92SNVDD1006 pKa = 3.88FDD1008 pKa = 5.56AFTDD1012 pKa = 3.85PFGADD1017 pKa = 3.01VSLGTGEE1024 pKa = 4.21SLKK1027 pKa = 10.89GRR1029 pKa = 11.84LGLSADD1035 pKa = 4.36YY1036 pKa = 11.05EE1037 pKa = 4.51NAWEE1041 pKa = 4.45GASGRR1046 pKa = 11.84TTRR1049 pKa = 11.84THH1051 pKa = 7.25LYY1053 pKa = 10.65GIANLYY1059 pKa = 10.82YY1060 pKa = 10.05EE1061 pKa = 4.84FLDD1064 pKa = 3.93GTEE1067 pKa = 4.22VEE1069 pKa = 4.39VSGVNFASEE1078 pKa = 4.2NDD1080 pKa = 3.97RR1081 pKa = 11.84TWGGIGTGGSYY1092 pKa = 10.33NWNDD1096 pKa = 3.7DD1097 pKa = 3.21KK1098 pKa = 11.77YY1099 pKa = 10.78SIYY1102 pKa = 10.97SEE1104 pKa = 4.38VSVNTSLSNFADD1116 pKa = 3.78SYY1118 pKa = 10.87SLNGTAGFRR1127 pKa = 11.84VKK1129 pKa = 10.46FF1130 pKa = 3.8
MM1 pKa = 7.39APAVAFADD9 pKa = 4.18CVTLGGVITCDD20 pKa = 3.03TAAPNPFTSTVGAGNTAAADD40 pKa = 3.98NITVNVGPGAQVAVGNLNAISLRR63 pKa = 11.84NNAVITIAGGGLVSATATSTTGNYY87 pKa = 7.37NTGGNAVEE95 pKa = 4.39VNNNGTLTIEE105 pKa = 4.4QGGQLLALGTQGSAEE120 pKa = 3.82AVNFQGTGNVLTNNGTIDD138 pKa = 3.65ANNSVAIWSQNTSGLNTVINNEE160 pKa = 4.0TGIIEE165 pKa = 4.59AGNGTTSTVIGGSGAGALDD184 pKa = 3.52FTNRR188 pKa = 11.84GTIRR192 pKa = 11.84GSINLANGNDD202 pKa = 3.4ILRR205 pKa = 11.84LYY207 pKa = 9.46TGSTVTGNFSGGGGTDD223 pKa = 4.04AIFLSGDD230 pKa = 3.77GQSSMAGNFNGFEE243 pKa = 4.18SLVKK247 pKa = 10.44NDD249 pKa = 3.7LGTWTLTGTITGVTVATVQNGTLVLTGANTNYY281 pKa = 9.01TGQVIIDD288 pKa = 3.6PAGTLEE294 pKa = 4.5ARR296 pKa = 11.84AQALPTQLLPANNVANITNNGLVRR320 pKa = 11.84FAQPDD325 pKa = 3.12NGTYY329 pKa = 10.03VGQIVGTGAVEE340 pKa = 3.87KK341 pKa = 9.83TGAGTLTLEE350 pKa = 4.39PSVAAGNTYY359 pKa = 10.78SGGTFINGGVVAVGADD375 pKa = 3.45NALGASTGGVSFNGGTLQLNSSFNLSAGRR404 pKa = 11.84AVTLNVGGGTINTQAFDD421 pKa = 3.38STLAQGVTGPGGLTKK436 pKa = 10.68AGAGTLTLTGNSDD449 pKa = 3.54YY450 pKa = 11.64SGGTVISAGTLQLGDD465 pKa = 4.02SGTSGSISGDD475 pKa = 3.33VLNNSLLAFNRR486 pKa = 11.84SDD488 pKa = 3.23AFLVPGVISGSGAVEE503 pKa = 3.92QNGSGTTILTGNNTYY518 pKa = 10.94SGGTTINAGTLQLGNGGTSGSIVGDD543 pKa = 3.6VANDD547 pKa = 3.39GSLAFNRR554 pKa = 11.84SDD556 pKa = 3.16STNFDD561 pKa = 3.59GVISSAGNVRR571 pKa = 11.84QIGGGTTILTAQNTYY586 pKa = 10.87SGATTVEE593 pKa = 4.03AGTLAAGATNVFSPNSSTSVLTGGTLDD620 pKa = 4.32AAGFDD625 pKa = 3.77QTVSALTNAGLVNVGGAPGTTLTVSGNYY653 pKa = 9.6VGADD657 pKa = 3.05GTLRR661 pKa = 11.84LNTALGDD668 pKa = 4.04DD669 pKa = 3.94ASTTDD674 pKa = 3.42RR675 pKa = 11.84LVVLGDD681 pKa = 3.67TSGSTTLVVANVGGSGAPTVEE702 pKa = 4.91GIRR705 pKa = 11.84VIDD708 pKa = 3.73VAGSSAGSFALDD720 pKa = 2.68GDD722 pKa = 4.33YY723 pKa = 11.36AFEE726 pKa = 4.8GDD728 pKa = 3.52EE729 pKa = 4.03AVIGGAYY736 pKa = 10.14AYY738 pKa = 10.24RR739 pKa = 11.84LYY741 pKa = 11.03KK742 pKa = 10.63NGIATPTDD750 pKa = 3.69GNWYY754 pKa = 9.8LRR756 pKa = 11.84SALVDD761 pKa = 3.64PADD764 pKa = 3.86PTAPTDD770 pKa = 3.67PTDD773 pKa = 3.87PTDD776 pKa = 3.89PTDD779 pKa = 3.76PTSPSSPLYY788 pKa = 9.64QPGVPVYY795 pKa = 9.58EE796 pKa = 5.31AYY798 pKa = 10.36AQTLLGLNGLPTLQQRR814 pKa = 11.84VGNRR818 pKa = 11.84YY819 pKa = 8.54WSGAGNGMLSQGDD832 pKa = 4.11GPGTIEE838 pKa = 4.97PAPQPSDD845 pKa = 3.12GGPAFVEE852 pKa = 5.1GRR854 pKa = 11.84GLWARR859 pKa = 11.84IEE861 pKa = 4.2GAHH864 pKa = 6.62GNFDD868 pKa = 3.87PRR870 pKa = 11.84TSTSSTDD877 pKa = 3.23YY878 pKa = 11.57DD879 pKa = 4.26LDD881 pKa = 3.3TWKK884 pKa = 10.27IQSGLDD890 pKa = 3.37GQLYY894 pKa = 9.89EE895 pKa = 4.52SEE897 pKa = 4.19SGKK900 pKa = 10.92LIGGITAQYY909 pKa = 10.11GNASADD915 pKa = 3.5VSSLFGNGEE924 pKa = 4.02VDD926 pKa = 2.95TDD928 pKa = 4.33GYY930 pKa = 11.6GVGATLTWYY939 pKa = 9.28GQNGFYY945 pKa = 10.78LDD947 pKa = 4.01GQAQATWYY955 pKa = 10.24EE956 pKa = 4.13SDD958 pKa = 3.81LSSGVLGSLSNGNDD972 pKa = 3.04GFGYY976 pKa = 10.62AFSLEE981 pKa = 4.2TGKK984 pKa = 10.72RR985 pKa = 11.84LALNQNWTLTPQAQLAYY1002 pKa = 10.92SNVDD1006 pKa = 3.88FDD1008 pKa = 5.56AFTDD1012 pKa = 3.85PFGADD1017 pKa = 3.01VSLGTGEE1024 pKa = 4.21SLKK1027 pKa = 10.89GRR1029 pKa = 11.84LGLSADD1035 pKa = 4.36YY1036 pKa = 11.05EE1037 pKa = 4.51NAWEE1041 pKa = 4.45GASGRR1046 pKa = 11.84TTRR1049 pKa = 11.84THH1051 pKa = 7.25LYY1053 pKa = 10.65GIANLYY1059 pKa = 10.82YY1060 pKa = 10.05EE1061 pKa = 4.84FLDD1064 pKa = 3.93GTEE1067 pKa = 4.22VEE1069 pKa = 4.39VSGVNFASEE1078 pKa = 4.2NDD1080 pKa = 3.97RR1081 pKa = 11.84TWGGIGTGGSYY1092 pKa = 10.33NWNDD1096 pKa = 3.7DD1097 pKa = 3.21KK1098 pKa = 11.77YY1099 pKa = 10.78SIYY1102 pKa = 10.97SEE1104 pKa = 4.38VSVNTSLSNFADD1116 pKa = 3.78SYY1118 pKa = 10.87SLNGTAGFRR1127 pKa = 11.84VKK1129 pKa = 10.46FF1130 pKa = 3.8
Molecular weight: 113.34 kDa
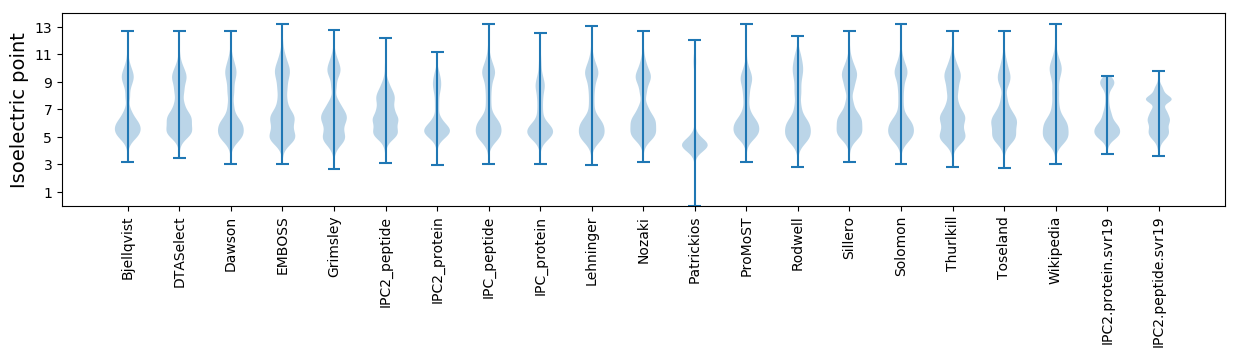
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N9VSU7|A0A2N9VSU7_9RHIZ YkuD domain-containing protein OS=Phyllobacterium zundukense OX=1867719 GN=B5P45_23120 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 9.15VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 9.15VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1679408 |
23 |
3389 |
299.6 |
32.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.314 ± 0.041 | 0.802 ± 0.009 |
5.673 ± 0.029 | 5.724 ± 0.033 |
3.976 ± 0.022 | 8.203 ± 0.029 |
2.077 ± 0.015 | 5.99 ± 0.021 |
4.002 ± 0.029 | 9.87 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.511 ± 0.016 | 3.072 ± 0.02 |
4.842 ± 0.025 | 3.167 ± 0.017 |
6.402 ± 0.028 | 5.84 ± 0.023 |
5.491 ± 0.027 | 7.341 ± 0.028 |
1.297 ± 0.012 | 2.403 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
