
Alternaria alternata fusarivirus 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.83
Get precalculated fractions of proteins
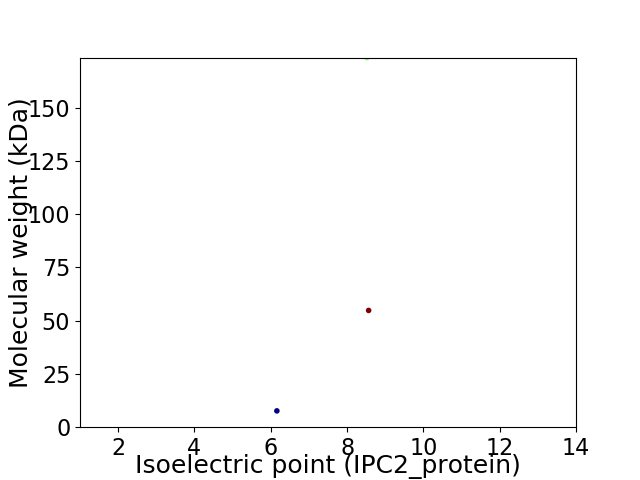
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z6DR81|A0A2Z6DR81_9VIRU Uncharacterized protein OS=Alternaria alternata fusarivirus 1 OX=1935440 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.26LVLLIRR8 pKa = 11.84PNPEE12 pKa = 3.22EE13 pKa = 4.08EE14 pKa = 4.15EE15 pKa = 4.3AVGSRR20 pKa = 11.84WCWKK24 pKa = 9.45HH25 pKa = 5.31PPYY28 pKa = 10.8VLSLSGTGFMSVRR41 pKa = 11.84GGGRR45 pKa = 11.84CAVRR49 pKa = 11.84AEE51 pKa = 4.09MFGNPEE57 pKa = 4.23CNDD60 pKa = 3.46CEE62 pKa = 4.51LFTRR66 pKa = 11.84FSS68 pKa = 3.37
MM1 pKa = 7.48KK2 pKa = 10.26LVLLIRR8 pKa = 11.84PNPEE12 pKa = 3.22EE13 pKa = 4.08EE14 pKa = 4.15EE15 pKa = 4.3AVGSRR20 pKa = 11.84WCWKK24 pKa = 9.45HH25 pKa = 5.31PPYY28 pKa = 10.8VLSLSGTGFMSVRR41 pKa = 11.84GGGRR45 pKa = 11.84CAVRR49 pKa = 11.84AEE51 pKa = 4.09MFGNPEE57 pKa = 4.23CNDD60 pKa = 3.46CEE62 pKa = 4.51LFTRR66 pKa = 11.84FSS68 pKa = 3.37
Molecular weight: 7.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z6DR81|A0A2Z6DR81_9VIRU Uncharacterized protein OS=Alternaria alternata fusarivirus 1 OX=1935440 PE=4 SV=1
MM1 pKa = 7.21TSEE4 pKa = 4.37KK5 pKa = 10.75VFIGKK10 pKa = 10.04RR11 pKa = 11.84IVQTRR16 pKa = 11.84EE17 pKa = 3.3QLSQVEE23 pKa = 4.1LTAQQVKK30 pKa = 10.47SSTVWLVRR38 pKa = 11.84EE39 pKa = 4.29PGWVSGQEE47 pKa = 4.03KK48 pKa = 10.43LNGWFLEE55 pKa = 4.26VEE57 pKa = 4.09GTDD60 pKa = 3.0IVLCVTTLFKK70 pKa = 11.58GEE72 pKa = 4.18VTSEE76 pKa = 3.5RR77 pKa = 11.84WFPPSPVFKK86 pKa = 10.0WKK88 pKa = 10.39KK89 pKa = 9.02EE90 pKa = 3.85YY91 pKa = 10.14QSRR94 pKa = 11.84DD95 pKa = 3.15KK96 pKa = 10.94EE97 pKa = 4.55VPTEE101 pKa = 3.81VAYY104 pKa = 10.2KK105 pKa = 9.68NVIVARR111 pKa = 11.84MADD114 pKa = 3.87EE115 pKa = 4.52NLLGYY120 pKa = 10.93YY121 pKa = 10.95DD122 pKa = 4.64MLTPATPLVNRR133 pKa = 11.84LSASFKK139 pKa = 8.0WWKK142 pKa = 8.96TGGNAPIIPFSPTNSKK158 pKa = 10.78KK159 pKa = 10.64EE160 pKa = 3.72IIGRR164 pKa = 11.84ISHH167 pKa = 7.03FEE169 pKa = 3.76TSFAALVNPPEE180 pKa = 4.12FALDD184 pKa = 3.3GLLALRR190 pKa = 11.84VLKK193 pKa = 10.71DD194 pKa = 3.4QLPSMKK200 pKa = 10.31DD201 pKa = 2.56RR202 pKa = 11.84GTQNVSRR209 pKa = 11.84FLTTLVKK216 pKa = 10.09AWAMSKK222 pKa = 10.14QPSHH226 pKa = 7.01FKK228 pKa = 11.08GPLSKK233 pKa = 9.93ISYY236 pKa = 8.93PVEE239 pKa = 4.42KK240 pKa = 10.46DD241 pKa = 3.14GLPADD246 pKa = 4.22RR247 pKa = 11.84AKK249 pKa = 10.07MYY251 pKa = 11.0HH252 pKa = 6.41NMICGKK258 pKa = 10.06GDD260 pKa = 3.84EE261 pKa = 4.36ITRR264 pKa = 11.84GLGFCMWTPPRR275 pKa = 11.84ITVTTDD281 pKa = 2.66AEE283 pKa = 4.69GFTTKK288 pKa = 9.8SRR290 pKa = 11.84KK291 pKa = 8.54KK292 pKa = 9.84HH293 pKa = 4.62EE294 pKa = 4.46EE295 pKa = 3.73KK296 pKa = 10.72KK297 pKa = 10.8IMLDD301 pKa = 3.97LGPDD305 pKa = 3.41IQVGGPNEE313 pKa = 3.8TRR315 pKa = 11.84LRR317 pKa = 11.84EE318 pKa = 3.92EE319 pKa = 4.41SEE321 pKa = 4.45AEE323 pKa = 4.06SSAMGARR330 pKa = 11.84RR331 pKa = 11.84QHH333 pKa = 5.91AKK335 pKa = 10.53KK336 pKa = 10.62NSDD339 pKa = 3.47PDD341 pKa = 3.54NGGFNPTSKK350 pKa = 10.35VSQKK354 pKa = 11.03DD355 pKa = 3.5QVSEE359 pKa = 4.22DD360 pKa = 3.88AVDD363 pKa = 5.64DD364 pKa = 3.94ATDD367 pKa = 3.19WLRR370 pKa = 11.84NSFTMTNFRR379 pKa = 11.84RR380 pKa = 11.84FYY382 pKa = 10.38RR383 pKa = 11.84RR384 pKa = 11.84VKK386 pKa = 10.49RR387 pKa = 11.84SVSSIKK393 pKa = 10.69DD394 pKa = 3.55SVTHH398 pKa = 6.19EE399 pKa = 4.08GVGIKK404 pKa = 10.38KK405 pKa = 10.07PFTEE409 pKa = 4.14KK410 pKa = 10.95AKK412 pKa = 10.48DD413 pKa = 3.4AVVFTVTSAVKK424 pKa = 10.34ACATVPFAGAMGLWYY439 pKa = 10.12GIKK442 pKa = 10.36EE443 pKa = 4.04PFVSIKK449 pKa = 10.2EE450 pKa = 4.05FPFPEE455 pKa = 4.49GRR457 pKa = 11.84LEE459 pKa = 4.24VAKK462 pKa = 10.58HH463 pKa = 4.96VADD466 pKa = 3.77CAVYY470 pKa = 10.1SLSSVVTSTISGVFAGLASATRR492 pKa = 3.68
MM1 pKa = 7.21TSEE4 pKa = 4.37KK5 pKa = 10.75VFIGKK10 pKa = 10.04RR11 pKa = 11.84IVQTRR16 pKa = 11.84EE17 pKa = 3.3QLSQVEE23 pKa = 4.1LTAQQVKK30 pKa = 10.47SSTVWLVRR38 pKa = 11.84EE39 pKa = 4.29PGWVSGQEE47 pKa = 4.03KK48 pKa = 10.43LNGWFLEE55 pKa = 4.26VEE57 pKa = 4.09GTDD60 pKa = 3.0IVLCVTTLFKK70 pKa = 11.58GEE72 pKa = 4.18VTSEE76 pKa = 3.5RR77 pKa = 11.84WFPPSPVFKK86 pKa = 10.0WKK88 pKa = 10.39KK89 pKa = 9.02EE90 pKa = 3.85YY91 pKa = 10.14QSRR94 pKa = 11.84DD95 pKa = 3.15KK96 pKa = 10.94EE97 pKa = 4.55VPTEE101 pKa = 3.81VAYY104 pKa = 10.2KK105 pKa = 9.68NVIVARR111 pKa = 11.84MADD114 pKa = 3.87EE115 pKa = 4.52NLLGYY120 pKa = 10.93YY121 pKa = 10.95DD122 pKa = 4.64MLTPATPLVNRR133 pKa = 11.84LSASFKK139 pKa = 8.0WWKK142 pKa = 8.96TGGNAPIIPFSPTNSKK158 pKa = 10.78KK159 pKa = 10.64EE160 pKa = 3.72IIGRR164 pKa = 11.84ISHH167 pKa = 7.03FEE169 pKa = 3.76TSFAALVNPPEE180 pKa = 4.12FALDD184 pKa = 3.3GLLALRR190 pKa = 11.84VLKK193 pKa = 10.71DD194 pKa = 3.4QLPSMKK200 pKa = 10.31DD201 pKa = 2.56RR202 pKa = 11.84GTQNVSRR209 pKa = 11.84FLTTLVKK216 pKa = 10.09AWAMSKK222 pKa = 10.14QPSHH226 pKa = 7.01FKK228 pKa = 11.08GPLSKK233 pKa = 9.93ISYY236 pKa = 8.93PVEE239 pKa = 4.42KK240 pKa = 10.46DD241 pKa = 3.14GLPADD246 pKa = 4.22RR247 pKa = 11.84AKK249 pKa = 10.07MYY251 pKa = 11.0HH252 pKa = 6.41NMICGKK258 pKa = 10.06GDD260 pKa = 3.84EE261 pKa = 4.36ITRR264 pKa = 11.84GLGFCMWTPPRR275 pKa = 11.84ITVTTDD281 pKa = 2.66AEE283 pKa = 4.69GFTTKK288 pKa = 9.8SRR290 pKa = 11.84KK291 pKa = 8.54KK292 pKa = 9.84HH293 pKa = 4.62EE294 pKa = 4.46EE295 pKa = 3.73KK296 pKa = 10.72KK297 pKa = 10.8IMLDD301 pKa = 3.97LGPDD305 pKa = 3.41IQVGGPNEE313 pKa = 3.8TRR315 pKa = 11.84LRR317 pKa = 11.84EE318 pKa = 3.92EE319 pKa = 4.41SEE321 pKa = 4.45AEE323 pKa = 4.06SSAMGARR330 pKa = 11.84RR331 pKa = 11.84QHH333 pKa = 5.91AKK335 pKa = 10.53KK336 pKa = 10.62NSDD339 pKa = 3.47PDD341 pKa = 3.54NGGFNPTSKK350 pKa = 10.35VSQKK354 pKa = 11.03DD355 pKa = 3.5QVSEE359 pKa = 4.22DD360 pKa = 3.88AVDD363 pKa = 5.64DD364 pKa = 3.94ATDD367 pKa = 3.19WLRR370 pKa = 11.84NSFTMTNFRR379 pKa = 11.84RR380 pKa = 11.84FYY382 pKa = 10.38RR383 pKa = 11.84RR384 pKa = 11.84VKK386 pKa = 10.49RR387 pKa = 11.84SVSSIKK393 pKa = 10.69DD394 pKa = 3.55SVTHH398 pKa = 6.19EE399 pKa = 4.08GVGIKK404 pKa = 10.38KK405 pKa = 10.07PFTEE409 pKa = 4.14KK410 pKa = 10.95AKK412 pKa = 10.48DD413 pKa = 3.4AVVFTVTSAVKK424 pKa = 10.34ACATVPFAGAMGLWYY439 pKa = 10.12GIKK442 pKa = 10.36EE443 pKa = 4.04PFVSIKK449 pKa = 10.2EE450 pKa = 4.05FPFPEE455 pKa = 4.49GRR457 pKa = 11.84LEE459 pKa = 4.24VAKK462 pKa = 10.58HH463 pKa = 4.96VADD466 pKa = 3.77CAVYY470 pKa = 10.1SLSSVVTSTISGVFAGLASATRR492 pKa = 3.68
Molecular weight: 54.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2082 |
68 |
1522 |
694.0 |
78.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.676 ± 0.289 | 0.768 ± 0.715 |
4.947 ± 0.515 | 6.052 ± 0.735 |
5.235 ± 0.089 | 5.956 ± 0.781 |
1.873 ± 0.247 | 4.947 ± 0.736 |
6.532 ± 0.919 | 9.99 ± 1.786 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.738 ± 0.21 | 3.65 ± 0.275 |
4.803 ± 0.641 | 2.546 ± 0.304 |
6.196 ± 0.382 | 7.589 ± 0.357 |
6.724 ± 0.495 | 7.541 ± 0.584 |
2.45 ± 0.1 | 2.786 ± 0.56 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
