
Drosophila busckii (Fruit fly)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera; Brachycera; Muscomorpha; Eremoneura; Cyclorrhapha; Schizophora; Acalyptrat
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
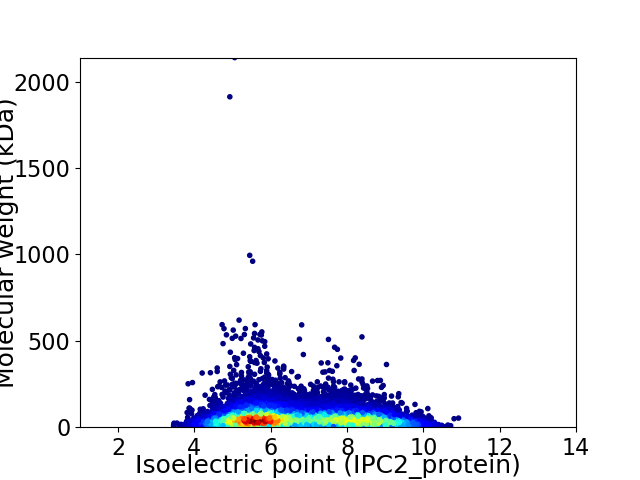
Virtual 2D-PAGE plot for 11914 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M4EM95|A0A0M4EM95_DROBS CG8500 OS=Drosophila busckii OX=30019 GN=Dbus_chr3Rg1290 PE=4 SV=1
MM1 pKa = 7.87ANLQADD7 pKa = 4.12KK8 pKa = 11.63SNGNCVYY15 pKa = 11.01ADD17 pKa = 4.02SCCHH21 pKa = 6.54TDD23 pKa = 3.22NAVATTATATSTTQNTTNTINIMTGAAAAAAVAASLPTSASCEE66 pKa = 4.08DD67 pKa = 4.08LDD69 pKa = 5.51LDD71 pKa = 4.12HH72 pKa = 6.77SQSLSEE78 pKa = 4.1YY79 pKa = 8.64TDD81 pKa = 3.56ADD83 pKa = 3.78EE84 pKa = 5.73SISAPTEE91 pKa = 3.66FLAEE95 pKa = 3.95FLSAVMLKK103 pKa = 10.31DD104 pKa = 3.34YY105 pKa = 11.09KK106 pKa = 10.76KK107 pKa = 10.48ALKK110 pKa = 9.48YY111 pKa = 10.51CKK113 pKa = 10.19LILQYY118 pKa = 10.82EE119 pKa = 4.47PNNTTAKK126 pKa = 10.05EE127 pKa = 4.1FYY129 pKa = 10.33PLILDD134 pKa = 3.91KK135 pKa = 11.29LRR137 pKa = 11.84VAAASDD143 pKa = 3.5SDD145 pKa = 3.65EE146 pKa = 4.72NYY148 pKa = 10.5NKK150 pKa = 10.42SSSPEE155 pKa = 3.82LNLEE159 pKa = 4.35LEE161 pKa = 4.56LEE163 pKa = 4.25AAADD167 pKa = 3.92CEE169 pKa = 4.22EE170 pKa = 4.85EE171 pKa = 4.1EE172 pKa = 5.57DD173 pKa = 4.16SVSSNSSDD181 pKa = 3.46ADD183 pKa = 3.52NDD185 pKa = 4.18DD186 pKa = 4.05EE187 pKa = 5.55VDD189 pKa = 4.23AEE191 pKa = 4.37PEE193 pKa = 3.98DD194 pKa = 4.33EE195 pKa = 4.64EE196 pKa = 4.68EE197 pKa = 4.38SSSGDD202 pKa = 3.82DD203 pKa = 3.44YY204 pKa = 11.54EE205 pKa = 5.59LPIDD209 pKa = 4.16EE210 pKa = 5.13PQDD213 pKa = 3.56VAAVGSLAAHH223 pKa = 5.51SHH225 pKa = 5.74SLSHH229 pKa = 6.63HH230 pKa = 6.11NSGSSRR236 pKa = 11.84NSSSGSGGRR245 pKa = 11.84SDD247 pKa = 3.93NTTHH251 pKa = 6.84SYY253 pKa = 11.27SSLLLEE259 pKa = 5.32DD260 pKa = 5.51DD261 pKa = 3.97NDD263 pKa = 3.79IVPLNLQFGNKK274 pKa = 8.87DD275 pKa = 3.18CHH277 pKa = 6.95AADD280 pKa = 4.37LDD282 pKa = 3.96VSNGNKK288 pKa = 9.39LASASCSDD296 pKa = 3.94SEE298 pKa = 5.32SPTEE302 pKa = 3.98PLTQRR307 pKa = 11.84LAAILRR313 pKa = 11.84SKK315 pKa = 10.79CGVSASNDD323 pKa = 3.23DD324 pKa = 3.77NYY326 pKa = 11.98
MM1 pKa = 7.87ANLQADD7 pKa = 4.12KK8 pKa = 11.63SNGNCVYY15 pKa = 11.01ADD17 pKa = 4.02SCCHH21 pKa = 6.54TDD23 pKa = 3.22NAVATTATATSTTQNTTNTINIMTGAAAAAAVAASLPTSASCEE66 pKa = 4.08DD67 pKa = 4.08LDD69 pKa = 5.51LDD71 pKa = 4.12HH72 pKa = 6.77SQSLSEE78 pKa = 4.1YY79 pKa = 8.64TDD81 pKa = 3.56ADD83 pKa = 3.78EE84 pKa = 5.73SISAPTEE91 pKa = 3.66FLAEE95 pKa = 3.95FLSAVMLKK103 pKa = 10.31DD104 pKa = 3.34YY105 pKa = 11.09KK106 pKa = 10.76KK107 pKa = 10.48ALKK110 pKa = 9.48YY111 pKa = 10.51CKK113 pKa = 10.19LILQYY118 pKa = 10.82EE119 pKa = 4.47PNNTTAKK126 pKa = 10.05EE127 pKa = 4.1FYY129 pKa = 10.33PLILDD134 pKa = 3.91KK135 pKa = 11.29LRR137 pKa = 11.84VAAASDD143 pKa = 3.5SDD145 pKa = 3.65EE146 pKa = 4.72NYY148 pKa = 10.5NKK150 pKa = 10.42SSSPEE155 pKa = 3.82LNLEE159 pKa = 4.35LEE161 pKa = 4.56LEE163 pKa = 4.25AAADD167 pKa = 3.92CEE169 pKa = 4.22EE170 pKa = 4.85EE171 pKa = 4.1EE172 pKa = 5.57DD173 pKa = 4.16SVSSNSSDD181 pKa = 3.46ADD183 pKa = 3.52NDD185 pKa = 4.18DD186 pKa = 4.05EE187 pKa = 5.55VDD189 pKa = 4.23AEE191 pKa = 4.37PEE193 pKa = 3.98DD194 pKa = 4.33EE195 pKa = 4.64EE196 pKa = 4.68EE197 pKa = 4.38SSSGDD202 pKa = 3.82DD203 pKa = 3.44YY204 pKa = 11.54EE205 pKa = 5.59LPIDD209 pKa = 4.16EE210 pKa = 5.13PQDD213 pKa = 3.56VAAVGSLAAHH223 pKa = 5.51SHH225 pKa = 5.74SLSHH229 pKa = 6.63HH230 pKa = 6.11NSGSSRR236 pKa = 11.84NSSSGSGGRR245 pKa = 11.84SDD247 pKa = 3.93NTTHH251 pKa = 6.84SYY253 pKa = 11.27SSLLLEE259 pKa = 5.32DD260 pKa = 5.51DD261 pKa = 3.97NDD263 pKa = 3.79IVPLNLQFGNKK274 pKa = 8.87DD275 pKa = 3.18CHH277 pKa = 6.95AADD280 pKa = 4.37LDD282 pKa = 3.96VSNGNKK288 pKa = 9.39LASASCSDD296 pKa = 3.94SEE298 pKa = 5.32SPTEE302 pKa = 3.98PLTQRR307 pKa = 11.84LAAILRR313 pKa = 11.84SKK315 pKa = 10.79CGVSASNDD323 pKa = 3.23DD324 pKa = 3.77NYY326 pKa = 11.98
Molecular weight: 34.51 kDa
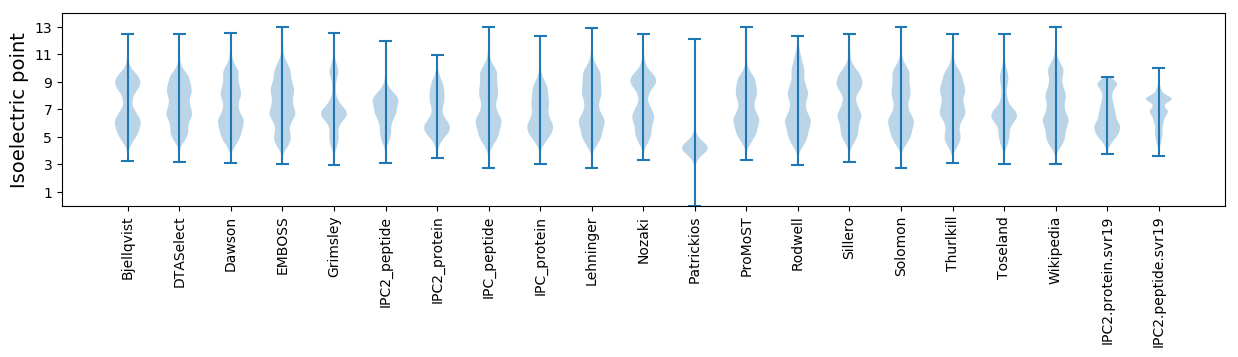
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M4F6C3|A0A0M4F6C3_DROBS Cyp6d5 OS=Drosophila busckii OX=30019 GN=Dbus_chr3Rg1982 PE=3 SV=1
VV1 pKa = 4.81QTGISLNDD9 pKa = 4.16RR10 pKa = 11.84FTQMQSRR17 pKa = 11.84QQQTAEE23 pKa = 3.82TRR25 pKa = 11.84GRR27 pKa = 11.84RR28 pKa = 11.84SRR30 pKa = 11.84SRR32 pKa = 11.84SRR34 pKa = 11.84SRR36 pKa = 11.84RR37 pKa = 11.84IVDD40 pKa = 3.51SAPSVANTRR49 pKa = 11.84LLQEE53 pKa = 5.16FKK55 pKa = 10.51RR56 pKa = 11.84KK57 pKa = 8.51HH58 pKa = 4.68TVQTALKK65 pKa = 9.27LKK67 pKa = 10.04RR68 pKa = 11.84RR69 pKa = 11.84SLRR72 pKa = 11.84TSASGASGMKK82 pKa = 10.23ALRR85 pKa = 11.84LGPNGKK91 pKa = 8.57PVRR94 pKa = 11.84SSNISRR100 pKa = 11.84LATMRR105 pKa = 11.84ADD107 pKa = 3.83LVASNGRR114 pKa = 11.84ARR116 pKa = 11.84RR117 pKa = 11.84SGSTTRR123 pKa = 11.84RR124 pKa = 11.84SNSAVRR130 pKa = 11.84GLRR133 pKa = 11.84QRR135 pKa = 11.84LGNRR139 pKa = 11.84RR140 pKa = 11.84PTVGGAAEE148 pKa = 3.98RR149 pKa = 11.84VAQRR153 pKa = 11.84RR154 pKa = 11.84RR155 pKa = 11.84GSLGRR160 pKa = 11.84FQQQQQQQQVLAPARR175 pKa = 11.84SRR177 pKa = 11.84SRR179 pKa = 11.84SRR181 pKa = 11.84ARR183 pKa = 11.84NTLSQPRR190 pKa = 11.84GRR192 pKa = 11.84ALSQPRR198 pKa = 11.84GRR200 pKa = 11.84PNSQVRR206 pKa = 11.84GRR208 pKa = 11.84EE209 pKa = 3.68RR210 pKa = 11.84SLSRR214 pKa = 11.84GRR216 pKa = 11.84DD217 pKa = 2.95RR218 pKa = 11.84SMGRR222 pKa = 11.84MPEE225 pKa = 3.86RR226 pKa = 11.84SQPRR230 pKa = 11.84ARR232 pKa = 11.84SQSRR236 pKa = 11.84GRR238 pKa = 11.84PPQVANSRR246 pKa = 11.84VSVKK250 pKa = 9.86QRR252 pKa = 11.84LGVRR256 pKa = 11.84PGVAATVAAAKK267 pKa = 9.85KK268 pKa = 9.88RR269 pKa = 11.84RR270 pKa = 11.84GNRR273 pKa = 11.84RR274 pKa = 11.84NSGQLPGVAAGRR286 pKa = 11.84IEE288 pKa = 3.81RR289 pKa = 11.84RR290 pKa = 11.84RR291 pKa = 11.84NSNVAQKK298 pKa = 10.77AVATAAGGRR307 pKa = 11.84GRR309 pKa = 11.84KK310 pKa = 8.93GRR312 pKa = 11.84ARR314 pKa = 11.84GRR316 pKa = 11.84SAVRR320 pKa = 11.84AAANASAAAVATQRR334 pKa = 11.84SRR336 pKa = 11.84SRR338 pKa = 11.84SRR340 pKa = 11.84SRR342 pKa = 11.84SRR344 pKa = 11.84ARR346 pKa = 11.84QGNAAAAGGAAAGKK360 pKa = 10.19GAGQSRR366 pKa = 11.84GRR368 pKa = 11.84PRR370 pKa = 11.84GRR372 pKa = 11.84AGKK375 pKa = 9.04VVKK378 pKa = 9.31ATNKK382 pKa = 10.21NGNNKK387 pKa = 9.63NSNINKK393 pKa = 9.47NKK395 pKa = 8.32NTGAGKK401 pKa = 10.35ANQQQGRR408 pKa = 11.84RR409 pKa = 11.84GRR411 pKa = 11.84SRR413 pKa = 11.84VRR415 pKa = 11.84GGGAGKK421 pKa = 8.51PQRR424 pKa = 11.84KK425 pKa = 6.99EE426 pKa = 3.91VKK428 pKa = 10.35RR429 pKa = 11.84EE430 pKa = 3.96DD431 pKa = 3.57LDD433 pKa = 4.86KK434 pKa = 11.45EE435 pKa = 4.02LDD437 pKa = 3.51QYY439 pKa = 11.55MSTTKK444 pKa = 11.22GEE446 pKa = 3.74MDD448 pKa = 4.0YY449 pKa = 11.4LLKK452 pKa = 10.93
VV1 pKa = 4.81QTGISLNDD9 pKa = 4.16RR10 pKa = 11.84FTQMQSRR17 pKa = 11.84QQQTAEE23 pKa = 3.82TRR25 pKa = 11.84GRR27 pKa = 11.84RR28 pKa = 11.84SRR30 pKa = 11.84SRR32 pKa = 11.84SRR34 pKa = 11.84SRR36 pKa = 11.84RR37 pKa = 11.84IVDD40 pKa = 3.51SAPSVANTRR49 pKa = 11.84LLQEE53 pKa = 5.16FKK55 pKa = 10.51RR56 pKa = 11.84KK57 pKa = 8.51HH58 pKa = 4.68TVQTALKK65 pKa = 9.27LKK67 pKa = 10.04RR68 pKa = 11.84RR69 pKa = 11.84SLRR72 pKa = 11.84TSASGASGMKK82 pKa = 10.23ALRR85 pKa = 11.84LGPNGKK91 pKa = 8.57PVRR94 pKa = 11.84SSNISRR100 pKa = 11.84LATMRR105 pKa = 11.84ADD107 pKa = 3.83LVASNGRR114 pKa = 11.84ARR116 pKa = 11.84RR117 pKa = 11.84SGSTTRR123 pKa = 11.84RR124 pKa = 11.84SNSAVRR130 pKa = 11.84GLRR133 pKa = 11.84QRR135 pKa = 11.84LGNRR139 pKa = 11.84RR140 pKa = 11.84PTVGGAAEE148 pKa = 3.98RR149 pKa = 11.84VAQRR153 pKa = 11.84RR154 pKa = 11.84RR155 pKa = 11.84GSLGRR160 pKa = 11.84FQQQQQQQQVLAPARR175 pKa = 11.84SRR177 pKa = 11.84SRR179 pKa = 11.84SRR181 pKa = 11.84ARR183 pKa = 11.84NTLSQPRR190 pKa = 11.84GRR192 pKa = 11.84ALSQPRR198 pKa = 11.84GRR200 pKa = 11.84PNSQVRR206 pKa = 11.84GRR208 pKa = 11.84EE209 pKa = 3.68RR210 pKa = 11.84SLSRR214 pKa = 11.84GRR216 pKa = 11.84DD217 pKa = 2.95RR218 pKa = 11.84SMGRR222 pKa = 11.84MPEE225 pKa = 3.86RR226 pKa = 11.84SQPRR230 pKa = 11.84ARR232 pKa = 11.84SQSRR236 pKa = 11.84GRR238 pKa = 11.84PPQVANSRR246 pKa = 11.84VSVKK250 pKa = 9.86QRR252 pKa = 11.84LGVRR256 pKa = 11.84PGVAATVAAAKK267 pKa = 9.85KK268 pKa = 9.88RR269 pKa = 11.84RR270 pKa = 11.84GNRR273 pKa = 11.84RR274 pKa = 11.84NSGQLPGVAAGRR286 pKa = 11.84IEE288 pKa = 3.81RR289 pKa = 11.84RR290 pKa = 11.84RR291 pKa = 11.84NSNVAQKK298 pKa = 10.77AVATAAGGRR307 pKa = 11.84GRR309 pKa = 11.84KK310 pKa = 8.93GRR312 pKa = 11.84ARR314 pKa = 11.84GRR316 pKa = 11.84SAVRR320 pKa = 11.84AAANASAAAVATQRR334 pKa = 11.84SRR336 pKa = 11.84SRR338 pKa = 11.84SRR340 pKa = 11.84SRR342 pKa = 11.84SRR344 pKa = 11.84ARR346 pKa = 11.84QGNAAAAGGAAAGKK360 pKa = 10.19GAGQSRR366 pKa = 11.84GRR368 pKa = 11.84PRR370 pKa = 11.84GRR372 pKa = 11.84AGKK375 pKa = 9.04VVKK378 pKa = 9.31ATNKK382 pKa = 10.21NGNNKK387 pKa = 9.63NSNINKK393 pKa = 9.47NKK395 pKa = 8.32NTGAGKK401 pKa = 10.35ANQQQGRR408 pKa = 11.84RR409 pKa = 11.84GRR411 pKa = 11.84SRR413 pKa = 11.84VRR415 pKa = 11.84GGGAGKK421 pKa = 8.51PQRR424 pKa = 11.84KK425 pKa = 6.99EE426 pKa = 3.91VKK428 pKa = 10.35RR429 pKa = 11.84EE430 pKa = 3.96DD431 pKa = 3.57LDD433 pKa = 4.86KK434 pKa = 11.45EE435 pKa = 4.02LDD437 pKa = 3.51QYY439 pKa = 11.55MSTTKK444 pKa = 11.22GEE446 pKa = 3.74MDD448 pKa = 4.0YY449 pKa = 11.4LLKK452 pKa = 10.93
Molecular weight: 49.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6422999 |
8 |
18943 |
539.1 |
60.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.149 ± 0.029 | 1.976 ± 0.019 |
5.053 ± 0.02 | 6.312 ± 0.033 |
3.508 ± 0.015 | 5.454 ± 0.026 |
2.666 ± 0.012 | 4.87 ± 0.019 |
5.493 ± 0.025 | 9.665 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.37 ± 0.01 | 4.861 ± 0.016 |
5.094 ± 0.024 | 5.644 ± 0.032 |
5.426 ± 0.021 | 7.91 ± 0.032 |
5.708 ± 0.022 | 5.681 ± 0.019 |
1.018 ± 0.007 | 3.141 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
