
Maribacter sp. T28
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Maribacter
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
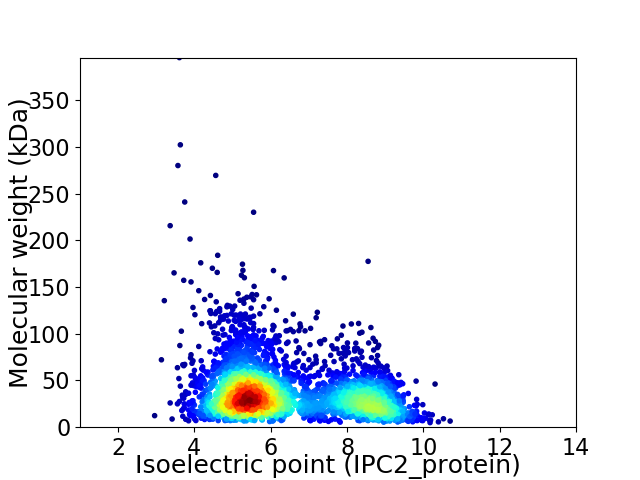
Virtual 2D-PAGE plot for 3578 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
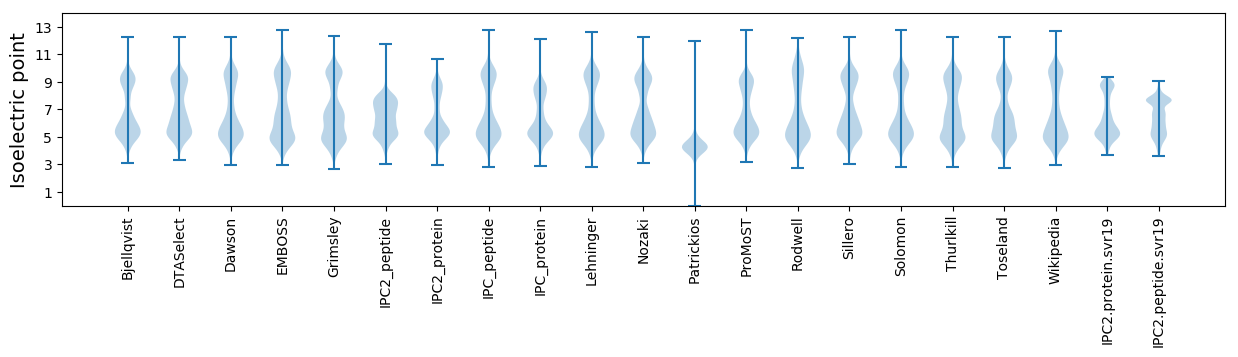
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B7Z8K2|A0A1B7Z8K2_9FLAO Rhomboid family intramembrane serine protease OS=Maribacter sp. T28 OX=1836467 GN=A9200_04285 PE=4 SV=1
MM1 pKa = 7.48RR2 pKa = 11.84TLLSCILFLCITLSYY17 pKa = 9.77SQSEE21 pKa = 5.41FITTWKK27 pKa = 8.84TDD29 pKa = 3.68NPGSTAPNSIVIPTYY44 pKa = 10.09PGEE47 pKa = 4.4TYY49 pKa = 10.37DD50 pKa = 4.06YY51 pKa = 10.4TVDD54 pKa = 3.32WGDD57 pKa = 3.46GTIEE61 pKa = 5.27SNLTDD66 pKa = 3.94NAIHH70 pKa = 6.77TYY72 pKa = 7.79ATPGIYY78 pKa = 9.97SVAISGTFPRR88 pKa = 11.84IYY90 pKa = 10.41FNNSGDD96 pKa = 3.78KK97 pKa = 10.7EE98 pKa = 4.14KK99 pKa = 11.03LILVNQWGSTNWSSMEE115 pKa = 4.11KK116 pKa = 10.55AFSGCINLDD125 pKa = 3.41VVATDD130 pKa = 3.5TPNFLGVNDD139 pKa = 4.94LSYY142 pKa = 11.32LFNGCTSLVGNNSFSNWNTSNVNNMRR168 pKa = 11.84AVFTDD173 pKa = 3.24TDD175 pKa = 4.21SFNQDD180 pKa = 2.22IGSWDD185 pKa = 3.41TSEE188 pKa = 4.46VINMAEE194 pKa = 3.96LFKK197 pKa = 11.04NSKK200 pKa = 10.15SFNQDD205 pKa = 1.97IGNWNISSVTNLAATFNGAILFNQNIGNWDD235 pKa = 3.43VSSVEE240 pKa = 4.03VFLGTFANAQNFNQNIGDD258 pKa = 3.92WDD260 pKa = 3.75VSAATDD266 pKa = 3.59MAGMFSNAISFNQDD280 pKa = 1.79IGRR283 pKa = 11.84WDD285 pKa = 3.36VSKK288 pKa = 10.81VKK290 pKa = 10.72RR291 pKa = 11.84MINMFNFAISFNQNIGFWNVSNVTSMNSMFYY322 pKa = 10.4NAQNFNQTLSYY333 pKa = 9.95WNVSNVEE340 pKa = 4.1NMNSMFDD347 pKa = 3.26RR348 pKa = 11.84ASNFDD353 pKa = 3.41QNLGYY358 pKa = 10.4WNISKK363 pKa = 8.23VTDD366 pKa = 3.82LEE368 pKa = 4.59YY369 pKa = 10.51MFSNAGLSTYY379 pKa = 10.92NYY381 pKa = 10.05DD382 pKa = 3.22ATLNGWAQLPILQSNLTFDD401 pKa = 4.18AGNSRR406 pKa = 11.84YY407 pKa = 9.82CFSEE411 pKa = 3.86TSRR414 pKa = 11.84NKK416 pKa = 10.42LLNDD420 pKa = 4.15LSWIINDD427 pKa = 3.34AGKK430 pKa = 9.95NLDD433 pKa = 4.24CKK435 pKa = 10.86PEE437 pKa = 4.15LFTTLWKK444 pKa = 9.45TDD446 pKa = 3.32NSGSSGDD453 pKa = 3.77NQITIPTFSGEE464 pKa = 4.02AYY466 pKa = 10.38NYY468 pKa = 7.79TVYY471 pKa = 10.19WGDD474 pKa = 3.43GTVSSNVTGNITHH487 pKa = 7.39TYY489 pKa = 8.38YY490 pKa = 10.81EE491 pKa = 4.27PGAYY495 pKa = 8.8QVSIEE500 pKa = 4.17GAFPRR505 pKa = 11.84IYY507 pKa = 10.62FNEE510 pKa = 4.03TGDD513 pKa = 3.53RR514 pKa = 11.84EE515 pKa = 4.32KK516 pKa = 10.67ILEE519 pKa = 4.1VKK521 pKa = 9.8QWGDD525 pKa = 3.33VSWNSMEE532 pKa = 4.38SAFAGCSNLSVAANDD547 pKa = 3.65VPNLSNTTSLKK558 pKa = 10.73RR559 pKa = 11.84MFLNCGYY566 pKa = 10.73AFDD569 pKa = 4.21FTRR572 pKa = 11.84SRR574 pKa = 11.84EE575 pKa = 3.79YY576 pKa = 11.13RR577 pKa = 11.84NFNGIEE583 pKa = 4.17NFNSWDD589 pKa = 3.34VSTITDD595 pKa = 3.66MSNMFDD601 pKa = 3.42KK602 pKa = 11.22SSFNQDD608 pKa = 1.91ISQWNVSNVTDD619 pKa = 3.79ISYY622 pKa = 10.48IFYY625 pKa = 10.4SSPFNFEE632 pKa = 3.47VSNWDD637 pKa = 3.17VSSVVNMQGVFGSSNFNKK655 pKa = 10.13SVSSWDD661 pKa = 3.37VSNVTNMDD669 pKa = 3.37YY670 pKa = 10.66MFNSTNFDD678 pKa = 3.13QDD680 pKa = 2.88ISSWNVSNVTSMRR693 pKa = 11.84NMLSQGSFNQDD704 pKa = 2.0ISSWNVSNVLDD715 pKa = 3.85MSNIFDD721 pKa = 5.23DD722 pKa = 4.09NDD724 pKa = 3.45LSRR727 pKa = 11.84EE728 pKa = 4.09NYY730 pKa = 10.36DD731 pKa = 3.8KK732 pKa = 11.04ILIEE736 pKa = 4.19WSLLPNLQNGIQLGAKK752 pKa = 9.51DD753 pKa = 3.7VQFCLGKK760 pKa = 10.13DD761 pKa = 3.24ARR763 pKa = 11.84DD764 pKa = 3.36ILTTIHH770 pKa = 6.07NWNIIDD776 pKa = 3.51QGEE779 pKa = 4.24NCEE782 pKa = 4.1EE783 pKa = 3.78EE784 pKa = 4.72RR785 pKa = 11.84PFVTTWKK792 pKa = 8.9TDD794 pKa = 3.22NPGFSDD800 pKa = 3.6NNQITIPTNPNKK812 pKa = 10.17VYY814 pKa = 10.46LYY816 pKa = 10.76NVDD819 pKa = 3.38WGDD822 pKa = 3.43GTFDD826 pKa = 3.21ISVTKK831 pKa = 10.44DD832 pKa = 2.67ITHH835 pKa = 7.07TYY837 pKa = 9.49VEE839 pKa = 4.6PGTYY843 pKa = 9.08QVSISGRR850 pKa = 11.84FPHH853 pKa = 7.52IYY855 pKa = 10.11FNNSGEE861 pKa = 4.6DD862 pKa = 3.89YY863 pKa = 10.7PHH865 pKa = 6.41QAPKK869 pKa = 9.1NTSDD873 pKa = 3.1IEE875 pKa = 4.32KK876 pKa = 10.08IISIDD881 pKa = 3.11QWGTNRR887 pKa = 11.84WISMAFAFAGCSNLDD902 pKa = 3.49VLATDD907 pKa = 4.21IPDD910 pKa = 3.79FSKK913 pKa = 11.16DD914 pKa = 2.83IDD916 pKa = 4.07LSFMFYY922 pKa = 10.81GCSSLKK928 pKa = 10.69GNEE931 pKa = 4.27SMEE934 pKa = 4.12LWDD937 pKa = 5.25LSDD940 pKa = 3.35ITMSTPSMFSGAIQFNQSLGNWDD963 pKa = 3.48VSNIRR968 pKa = 11.84YY969 pKa = 9.04LDD971 pKa = 3.34RR972 pKa = 11.84MFEE975 pKa = 4.03NAISFNQNLEE985 pKa = 3.5NWDD988 pKa = 3.33IGKK991 pKa = 9.51VEE993 pKa = 4.49NMDD996 pKa = 3.76AMFNGSGISIANYY1009 pKa = 10.09DD1010 pKa = 3.84RR1011 pKa = 11.84LLNAWSTLPSLVNNVNLGAADD1032 pKa = 3.88TNFCNSTEE1040 pKa = 3.82ARR1042 pKa = 11.84QNLIDD1047 pKa = 3.65TYY1049 pKa = 10.94GWQITDD1055 pKa = 3.93AGNDD1059 pKa = 3.34CSGTYY1064 pKa = 10.43FITTWKK1070 pKa = 8.6TDD1072 pKa = 3.32NPGISADD1079 pKa = 3.77NQITIPTFPSEE1090 pKa = 4.26TYY1092 pKa = 10.02DD1093 pKa = 4.06YY1094 pKa = 11.18SVNWGDD1100 pKa = 5.26GNIEE1104 pKa = 4.59SNFIGDD1110 pKa = 3.56ATHH1113 pKa = 7.3TYY1115 pKa = 7.63LTPGTYY1121 pKa = 9.15QVSISGVFPRR1131 pKa = 11.84IFFNNFDD1138 pKa = 4.09GTSSDD1143 pKa = 3.04SDD1145 pKa = 3.84KK1146 pKa = 10.98IISVDD1151 pKa = 3.06QWGDD1155 pKa = 3.58IEE1157 pKa = 4.38WTSMNNAFTNCTNLNVVAIDD1177 pKa = 3.66IPNLEE1182 pKa = 4.43NVNSLGAMFRR1192 pKa = 11.84FCDD1195 pKa = 3.59SLIGNEE1201 pKa = 5.09SFSKK1205 pKa = 9.71WQVKK1209 pKa = 9.97SINFFDD1215 pKa = 4.21FLFDD1219 pKa = 3.63GASQFNQYY1227 pKa = 9.17IGNWDD1232 pKa = 3.43VSNATNMSYY1241 pKa = 9.69MFNNATIFDD1250 pKa = 3.87QDD1252 pKa = 3.04ITNWDD1257 pKa = 3.64ISNVDD1262 pKa = 3.69HH1263 pKa = 7.19IGLMFANTKK1272 pKa = 10.29AFNQPIGNWNMSNLTSLQAIFAGASAFNQDD1302 pKa = 2.5ISSWDD1307 pKa = 3.43VSNVTNMEE1315 pKa = 3.93GMFSGAISFNQDD1327 pKa = 1.81ISNWDD1332 pKa = 3.45VSNVTTMNSMFPGAISFNSPLNSWDD1357 pKa = 3.51VSSVTDD1363 pKa = 3.51MTAMFDD1369 pKa = 3.43GATSFNRR1376 pKa = 11.84PLNNWNISNVTQISSMFVNATSFNQPLDD1404 pKa = 3.32NWDD1407 pKa = 3.57VSNVTSLAGMFLKK1420 pKa = 10.75ASSFNQNLSNWDD1432 pKa = 3.23ISNVTSIGGMFNQAISYY1449 pKa = 8.31DD1450 pKa = 3.92QNLGDD1455 pKa = 3.87WDD1457 pKa = 3.74VSKK1460 pKa = 10.57IINMDD1465 pKa = 3.56YY1466 pKa = 10.75LFNGIGLSLDD1476 pKa = 4.16NYY1478 pKa = 11.16DD1479 pKa = 3.65KK1480 pKa = 11.2TLTGWANLPSLQNNVVFDD1498 pKa = 4.07AANSKK1503 pKa = 10.46YY1504 pKa = 10.41CEE1506 pKa = 4.26SEE1508 pKa = 3.77NARR1511 pKa = 11.84QSIIDD1516 pKa = 3.64TFGWTINDD1524 pKa = 3.39FGKK1527 pKa = 10.81VPFCNEE1533 pKa = 4.0DD1534 pKa = 3.26NDD1536 pKa = 4.4LDD1538 pKa = 5.31GILDD1542 pKa = 4.17HH1543 pKa = 7.43KK1544 pKa = 10.93DD1545 pKa = 3.03SCLDD1549 pKa = 3.16TRR1551 pKa = 11.84PNVTVNDD1558 pKa = 3.97NGCEE1562 pKa = 3.95IIVANAILVYY1572 pKa = 9.99GASPTCPGEE1581 pKa = 4.03ANGTIEE1587 pKa = 4.95ISSNLINYY1595 pKa = 7.49GFNISIEE1602 pKa = 4.38GPVSFDD1608 pKa = 3.39YY1609 pKa = 11.72NEE1611 pKa = 3.82ISLNNNLEE1619 pKa = 4.01VSNLTTGMYY1628 pKa = 8.83TVTIAIPDD1636 pKa = 3.35ISYY1639 pKa = 9.51VQTYY1643 pKa = 9.03GIQINEE1649 pKa = 3.93VDD1651 pKa = 3.98TISGKK1656 pKa = 10.19RR1657 pKa = 11.84EE1658 pKa = 3.87NLNTSAKK1665 pKa = 7.61TALYY1669 pKa = 9.99NVEE1672 pKa = 3.62GSYY1675 pKa = 11.17NYY1677 pKa = 9.7TVDD1680 pKa = 4.19VNGEE1684 pKa = 3.95LKK1686 pKa = 10.78NYY1688 pKa = 9.8NFTSNGVNEE1697 pKa = 4.41IQLSDD1702 pKa = 3.59LGEE1705 pKa = 4.3INTISISGEE1714 pKa = 3.72SDD1716 pKa = 3.44CQGKK1720 pKa = 7.96VTDD1723 pKa = 4.79SFAFTDD1729 pKa = 4.14GLIMYY1734 pKa = 7.35PTITTGEE1741 pKa = 4.3VFVEE1745 pKa = 4.89GCDD1748 pKa = 3.48EE1749 pKa = 4.3SSTVLVYY1756 pKa = 10.69DD1757 pKa = 3.61LSGRR1761 pKa = 11.84LVFSKK1766 pKa = 9.57TLSPQDD1772 pKa = 3.86SNSIDD1777 pKa = 3.23IQALEE1782 pKa = 4.14TGVYY1786 pKa = 10.27PMVIQSKK1793 pKa = 9.87EE1794 pKa = 3.57NSKK1797 pKa = 8.08TFKK1800 pKa = 10.33IIKK1803 pKa = 8.78QQ1804 pKa = 3.43
MM1 pKa = 7.48RR2 pKa = 11.84TLLSCILFLCITLSYY17 pKa = 9.77SQSEE21 pKa = 5.41FITTWKK27 pKa = 8.84TDD29 pKa = 3.68NPGSTAPNSIVIPTYY44 pKa = 10.09PGEE47 pKa = 4.4TYY49 pKa = 10.37DD50 pKa = 4.06YY51 pKa = 10.4TVDD54 pKa = 3.32WGDD57 pKa = 3.46GTIEE61 pKa = 5.27SNLTDD66 pKa = 3.94NAIHH70 pKa = 6.77TYY72 pKa = 7.79ATPGIYY78 pKa = 9.97SVAISGTFPRR88 pKa = 11.84IYY90 pKa = 10.41FNNSGDD96 pKa = 3.78KK97 pKa = 10.7EE98 pKa = 4.14KK99 pKa = 11.03LILVNQWGSTNWSSMEE115 pKa = 4.11KK116 pKa = 10.55AFSGCINLDD125 pKa = 3.41VVATDD130 pKa = 3.5TPNFLGVNDD139 pKa = 4.94LSYY142 pKa = 11.32LFNGCTSLVGNNSFSNWNTSNVNNMRR168 pKa = 11.84AVFTDD173 pKa = 3.24TDD175 pKa = 4.21SFNQDD180 pKa = 2.22IGSWDD185 pKa = 3.41TSEE188 pKa = 4.46VINMAEE194 pKa = 3.96LFKK197 pKa = 11.04NSKK200 pKa = 10.15SFNQDD205 pKa = 1.97IGNWNISSVTNLAATFNGAILFNQNIGNWDD235 pKa = 3.43VSSVEE240 pKa = 4.03VFLGTFANAQNFNQNIGDD258 pKa = 3.92WDD260 pKa = 3.75VSAATDD266 pKa = 3.59MAGMFSNAISFNQDD280 pKa = 1.79IGRR283 pKa = 11.84WDD285 pKa = 3.36VSKK288 pKa = 10.81VKK290 pKa = 10.72RR291 pKa = 11.84MINMFNFAISFNQNIGFWNVSNVTSMNSMFYY322 pKa = 10.4NAQNFNQTLSYY333 pKa = 9.95WNVSNVEE340 pKa = 4.1NMNSMFDD347 pKa = 3.26RR348 pKa = 11.84ASNFDD353 pKa = 3.41QNLGYY358 pKa = 10.4WNISKK363 pKa = 8.23VTDD366 pKa = 3.82LEE368 pKa = 4.59YY369 pKa = 10.51MFSNAGLSTYY379 pKa = 10.92NYY381 pKa = 10.05DD382 pKa = 3.22ATLNGWAQLPILQSNLTFDD401 pKa = 4.18AGNSRR406 pKa = 11.84YY407 pKa = 9.82CFSEE411 pKa = 3.86TSRR414 pKa = 11.84NKK416 pKa = 10.42LLNDD420 pKa = 4.15LSWIINDD427 pKa = 3.34AGKK430 pKa = 9.95NLDD433 pKa = 4.24CKK435 pKa = 10.86PEE437 pKa = 4.15LFTTLWKK444 pKa = 9.45TDD446 pKa = 3.32NSGSSGDD453 pKa = 3.77NQITIPTFSGEE464 pKa = 4.02AYY466 pKa = 10.38NYY468 pKa = 7.79TVYY471 pKa = 10.19WGDD474 pKa = 3.43GTVSSNVTGNITHH487 pKa = 7.39TYY489 pKa = 8.38YY490 pKa = 10.81EE491 pKa = 4.27PGAYY495 pKa = 8.8QVSIEE500 pKa = 4.17GAFPRR505 pKa = 11.84IYY507 pKa = 10.62FNEE510 pKa = 4.03TGDD513 pKa = 3.53RR514 pKa = 11.84EE515 pKa = 4.32KK516 pKa = 10.67ILEE519 pKa = 4.1VKK521 pKa = 9.8QWGDD525 pKa = 3.33VSWNSMEE532 pKa = 4.38SAFAGCSNLSVAANDD547 pKa = 3.65VPNLSNTTSLKK558 pKa = 10.73RR559 pKa = 11.84MFLNCGYY566 pKa = 10.73AFDD569 pKa = 4.21FTRR572 pKa = 11.84SRR574 pKa = 11.84EE575 pKa = 3.79YY576 pKa = 11.13RR577 pKa = 11.84NFNGIEE583 pKa = 4.17NFNSWDD589 pKa = 3.34VSTITDD595 pKa = 3.66MSNMFDD601 pKa = 3.42KK602 pKa = 11.22SSFNQDD608 pKa = 1.91ISQWNVSNVTDD619 pKa = 3.79ISYY622 pKa = 10.48IFYY625 pKa = 10.4SSPFNFEE632 pKa = 3.47VSNWDD637 pKa = 3.17VSSVVNMQGVFGSSNFNKK655 pKa = 10.13SVSSWDD661 pKa = 3.37VSNVTNMDD669 pKa = 3.37YY670 pKa = 10.66MFNSTNFDD678 pKa = 3.13QDD680 pKa = 2.88ISSWNVSNVTSMRR693 pKa = 11.84NMLSQGSFNQDD704 pKa = 2.0ISSWNVSNVLDD715 pKa = 3.85MSNIFDD721 pKa = 5.23DD722 pKa = 4.09NDD724 pKa = 3.45LSRR727 pKa = 11.84EE728 pKa = 4.09NYY730 pKa = 10.36DD731 pKa = 3.8KK732 pKa = 11.04ILIEE736 pKa = 4.19WSLLPNLQNGIQLGAKK752 pKa = 9.51DD753 pKa = 3.7VQFCLGKK760 pKa = 10.13DD761 pKa = 3.24ARR763 pKa = 11.84DD764 pKa = 3.36ILTTIHH770 pKa = 6.07NWNIIDD776 pKa = 3.51QGEE779 pKa = 4.24NCEE782 pKa = 4.1EE783 pKa = 3.78EE784 pKa = 4.72RR785 pKa = 11.84PFVTTWKK792 pKa = 8.9TDD794 pKa = 3.22NPGFSDD800 pKa = 3.6NNQITIPTNPNKK812 pKa = 10.17VYY814 pKa = 10.46LYY816 pKa = 10.76NVDD819 pKa = 3.38WGDD822 pKa = 3.43GTFDD826 pKa = 3.21ISVTKK831 pKa = 10.44DD832 pKa = 2.67ITHH835 pKa = 7.07TYY837 pKa = 9.49VEE839 pKa = 4.6PGTYY843 pKa = 9.08QVSISGRR850 pKa = 11.84FPHH853 pKa = 7.52IYY855 pKa = 10.11FNNSGEE861 pKa = 4.6DD862 pKa = 3.89YY863 pKa = 10.7PHH865 pKa = 6.41QAPKK869 pKa = 9.1NTSDD873 pKa = 3.1IEE875 pKa = 4.32KK876 pKa = 10.08IISIDD881 pKa = 3.11QWGTNRR887 pKa = 11.84WISMAFAFAGCSNLDD902 pKa = 3.49VLATDD907 pKa = 4.21IPDD910 pKa = 3.79FSKK913 pKa = 11.16DD914 pKa = 2.83IDD916 pKa = 4.07LSFMFYY922 pKa = 10.81GCSSLKK928 pKa = 10.69GNEE931 pKa = 4.27SMEE934 pKa = 4.12LWDD937 pKa = 5.25LSDD940 pKa = 3.35ITMSTPSMFSGAIQFNQSLGNWDD963 pKa = 3.48VSNIRR968 pKa = 11.84YY969 pKa = 9.04LDD971 pKa = 3.34RR972 pKa = 11.84MFEE975 pKa = 4.03NAISFNQNLEE985 pKa = 3.5NWDD988 pKa = 3.33IGKK991 pKa = 9.51VEE993 pKa = 4.49NMDD996 pKa = 3.76AMFNGSGISIANYY1009 pKa = 10.09DD1010 pKa = 3.84RR1011 pKa = 11.84LLNAWSTLPSLVNNVNLGAADD1032 pKa = 3.88TNFCNSTEE1040 pKa = 3.82ARR1042 pKa = 11.84QNLIDD1047 pKa = 3.65TYY1049 pKa = 10.94GWQITDD1055 pKa = 3.93AGNDD1059 pKa = 3.34CSGTYY1064 pKa = 10.43FITTWKK1070 pKa = 8.6TDD1072 pKa = 3.32NPGISADD1079 pKa = 3.77NQITIPTFPSEE1090 pKa = 4.26TYY1092 pKa = 10.02DD1093 pKa = 4.06YY1094 pKa = 11.18SVNWGDD1100 pKa = 5.26GNIEE1104 pKa = 4.59SNFIGDD1110 pKa = 3.56ATHH1113 pKa = 7.3TYY1115 pKa = 7.63LTPGTYY1121 pKa = 9.15QVSISGVFPRR1131 pKa = 11.84IFFNNFDD1138 pKa = 4.09GTSSDD1143 pKa = 3.04SDD1145 pKa = 3.84KK1146 pKa = 10.98IISVDD1151 pKa = 3.06QWGDD1155 pKa = 3.58IEE1157 pKa = 4.38WTSMNNAFTNCTNLNVVAIDD1177 pKa = 3.66IPNLEE1182 pKa = 4.43NVNSLGAMFRR1192 pKa = 11.84FCDD1195 pKa = 3.59SLIGNEE1201 pKa = 5.09SFSKK1205 pKa = 9.71WQVKK1209 pKa = 9.97SINFFDD1215 pKa = 4.21FLFDD1219 pKa = 3.63GASQFNQYY1227 pKa = 9.17IGNWDD1232 pKa = 3.43VSNATNMSYY1241 pKa = 9.69MFNNATIFDD1250 pKa = 3.87QDD1252 pKa = 3.04ITNWDD1257 pKa = 3.64ISNVDD1262 pKa = 3.69HH1263 pKa = 7.19IGLMFANTKK1272 pKa = 10.29AFNQPIGNWNMSNLTSLQAIFAGASAFNQDD1302 pKa = 2.5ISSWDD1307 pKa = 3.43VSNVTNMEE1315 pKa = 3.93GMFSGAISFNQDD1327 pKa = 1.81ISNWDD1332 pKa = 3.45VSNVTTMNSMFPGAISFNSPLNSWDD1357 pKa = 3.51VSSVTDD1363 pKa = 3.51MTAMFDD1369 pKa = 3.43GATSFNRR1376 pKa = 11.84PLNNWNISNVTQISSMFVNATSFNQPLDD1404 pKa = 3.32NWDD1407 pKa = 3.57VSNVTSLAGMFLKK1420 pKa = 10.75ASSFNQNLSNWDD1432 pKa = 3.23ISNVTSIGGMFNQAISYY1449 pKa = 8.31DD1450 pKa = 3.92QNLGDD1455 pKa = 3.87WDD1457 pKa = 3.74VSKK1460 pKa = 10.57IINMDD1465 pKa = 3.56YY1466 pKa = 10.75LFNGIGLSLDD1476 pKa = 4.16NYY1478 pKa = 11.16DD1479 pKa = 3.65KK1480 pKa = 11.2TLTGWANLPSLQNNVVFDD1498 pKa = 4.07AANSKK1503 pKa = 10.46YY1504 pKa = 10.41CEE1506 pKa = 4.26SEE1508 pKa = 3.77NARR1511 pKa = 11.84QSIIDD1516 pKa = 3.64TFGWTINDD1524 pKa = 3.39FGKK1527 pKa = 10.81VPFCNEE1533 pKa = 4.0DD1534 pKa = 3.26NDD1536 pKa = 4.4LDD1538 pKa = 5.31GILDD1542 pKa = 4.17HH1543 pKa = 7.43KK1544 pKa = 10.93DD1545 pKa = 3.03SCLDD1549 pKa = 3.16TRR1551 pKa = 11.84PNVTVNDD1558 pKa = 3.97NGCEE1562 pKa = 3.95IIVANAILVYY1572 pKa = 9.99GASPTCPGEE1581 pKa = 4.03ANGTIEE1587 pKa = 4.95ISSNLINYY1595 pKa = 7.49GFNISIEE1602 pKa = 4.38GPVSFDD1608 pKa = 3.39YY1609 pKa = 11.72NEE1611 pKa = 3.82ISLNNNLEE1619 pKa = 4.01VSNLTTGMYY1628 pKa = 8.83TVTIAIPDD1636 pKa = 3.35ISYY1639 pKa = 9.51VQTYY1643 pKa = 9.03GIQINEE1649 pKa = 3.93VDD1651 pKa = 3.98TISGKK1656 pKa = 10.19RR1657 pKa = 11.84EE1658 pKa = 3.87NLNTSAKK1665 pKa = 7.61TALYY1669 pKa = 9.99NVEE1672 pKa = 3.62GSYY1675 pKa = 11.17NYY1677 pKa = 9.7TVDD1680 pKa = 4.19VNGEE1684 pKa = 3.95LKK1686 pKa = 10.78NYY1688 pKa = 9.8NFTSNGVNEE1697 pKa = 4.41IQLSDD1702 pKa = 3.59LGEE1705 pKa = 4.3INTISISGEE1714 pKa = 3.72SDD1716 pKa = 3.44CQGKK1720 pKa = 7.96VTDD1723 pKa = 4.79SFAFTDD1729 pKa = 4.14GLIMYY1734 pKa = 7.35PTITTGEE1741 pKa = 4.3VFVEE1745 pKa = 4.89GCDD1748 pKa = 3.48EE1749 pKa = 4.3SSTVLVYY1756 pKa = 10.69DD1757 pKa = 3.61LSGRR1761 pKa = 11.84LVFSKK1766 pKa = 9.57TLSPQDD1772 pKa = 3.86SNSIDD1777 pKa = 3.23IQALEE1782 pKa = 4.14TGVYY1786 pKa = 10.27PMVIQSKK1793 pKa = 9.87EE1794 pKa = 3.57NSKK1797 pKa = 8.08TFKK1800 pKa = 10.33IIKK1803 pKa = 8.78QQ1804 pKa = 3.43
Molecular weight: 201.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B7Z1R9|A0A1B7Z1R9_9FLAO LPS export ABC transporter ATP-binding protein OS=Maribacter sp. T28 OX=1836467 GN=A9200_09685 PE=4 SV=1
MM1 pKa = 7.02YY2 pKa = 10.31LVRR5 pKa = 11.84KK6 pKa = 10.0INFTDD11 pKa = 2.98NGQRR15 pKa = 11.84TTDD18 pKa = 3.03NGQRR22 pKa = 11.84TTDD25 pKa = 3.03NGQRR29 pKa = 11.84TTDD32 pKa = 3.03NGQRR36 pKa = 11.84TTDD39 pKa = 3.03NGQRR43 pKa = 11.84TTDD46 pKa = 3.03NGQRR50 pKa = 11.84TTDD53 pKa = 3.03NGQRR57 pKa = 11.84TTDD60 pKa = 3.03NGQRR64 pKa = 11.84TTDD67 pKa = 3.03NGQRR71 pKa = 11.84TTDD74 pKa = 3.03NGQRR78 pKa = 11.84TTDD81 pKa = 3.03NGQRR85 pKa = 11.84TTDD88 pKa = 3.03NGQRR92 pKa = 11.84TTDD95 pKa = 3.03NGQRR99 pKa = 11.84TTDD102 pKa = 3.03NGQRR106 pKa = 11.84TTDD109 pKa = 3.03NGQRR113 pKa = 11.84TTDD116 pKa = 3.03NGQRR120 pKa = 11.84TTDD123 pKa = 3.03NGQRR127 pKa = 11.84TTDD130 pKa = 3.03NGQRR134 pKa = 11.84TTDD137 pKa = 3.03NGQRR141 pKa = 11.84TTDD144 pKa = 3.03NGQRR148 pKa = 11.84TTDD151 pKa = 3.03NGQRR155 pKa = 11.84TTDD158 pKa = 3.03NGQRR162 pKa = 11.84TTDD165 pKa = 3.03NGQRR169 pKa = 11.84TTDD172 pKa = 3.03NGQRR176 pKa = 11.84TTDD179 pKa = 3.03NGQRR183 pKa = 11.84TTDD186 pKa = 3.03NGQRR190 pKa = 11.84TTDD193 pKa = 3.03NGQRR197 pKa = 11.84TTDD200 pKa = 3.03NGQRR204 pKa = 11.84TTDD207 pKa = 3.03NGQRR211 pKa = 11.84TTDD214 pKa = 3.03NGQRR218 pKa = 11.84TTDD221 pKa = 3.03NGQRR225 pKa = 11.84KK226 pKa = 8.83ARR228 pKa = 11.84FNLKK232 pKa = 10.45LILNLRR238 pKa = 11.84LYY240 pKa = 10.82
MM1 pKa = 7.02YY2 pKa = 10.31LVRR5 pKa = 11.84KK6 pKa = 10.0INFTDD11 pKa = 2.98NGQRR15 pKa = 11.84TTDD18 pKa = 3.03NGQRR22 pKa = 11.84TTDD25 pKa = 3.03NGQRR29 pKa = 11.84TTDD32 pKa = 3.03NGQRR36 pKa = 11.84TTDD39 pKa = 3.03NGQRR43 pKa = 11.84TTDD46 pKa = 3.03NGQRR50 pKa = 11.84TTDD53 pKa = 3.03NGQRR57 pKa = 11.84TTDD60 pKa = 3.03NGQRR64 pKa = 11.84TTDD67 pKa = 3.03NGQRR71 pKa = 11.84TTDD74 pKa = 3.03NGQRR78 pKa = 11.84TTDD81 pKa = 3.03NGQRR85 pKa = 11.84TTDD88 pKa = 3.03NGQRR92 pKa = 11.84TTDD95 pKa = 3.03NGQRR99 pKa = 11.84TTDD102 pKa = 3.03NGQRR106 pKa = 11.84TTDD109 pKa = 3.03NGQRR113 pKa = 11.84TTDD116 pKa = 3.03NGQRR120 pKa = 11.84TTDD123 pKa = 3.03NGQRR127 pKa = 11.84TTDD130 pKa = 3.03NGQRR134 pKa = 11.84TTDD137 pKa = 3.03NGQRR141 pKa = 11.84TTDD144 pKa = 3.03NGQRR148 pKa = 11.84TTDD151 pKa = 3.03NGQRR155 pKa = 11.84TTDD158 pKa = 3.03NGQRR162 pKa = 11.84TTDD165 pKa = 3.03NGQRR169 pKa = 11.84TTDD172 pKa = 3.03NGQRR176 pKa = 11.84TTDD179 pKa = 3.03NGQRR183 pKa = 11.84TTDD186 pKa = 3.03NGQRR190 pKa = 11.84TTDD193 pKa = 3.03NGQRR197 pKa = 11.84TTDD200 pKa = 3.03NGQRR204 pKa = 11.84TTDD207 pKa = 3.03NGQRR211 pKa = 11.84TTDD214 pKa = 3.03NGQRR218 pKa = 11.84TTDD221 pKa = 3.03NGQRR225 pKa = 11.84KK226 pKa = 8.83ARR228 pKa = 11.84FNLKK232 pKa = 10.45LILNLRR238 pKa = 11.84LYY240 pKa = 10.82
Molecular weight: 26.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1244144 |
42 |
3735 |
347.7 |
39.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.336 ± 0.035 | 0.71 ± 0.013 |
5.661 ± 0.033 | 6.676 ± 0.03 |
5.157 ± 0.031 | 6.572 ± 0.041 |
1.723 ± 0.018 | 7.978 ± 0.045 |
7.654 ± 0.053 | 9.418 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.286 ± 0.023 | 6.226 ± 0.045 |
3.418 ± 0.022 | 3.215 ± 0.02 |
3.319 ± 0.023 | 6.536 ± 0.03 |
5.837 ± 0.045 | 6.197 ± 0.033 |
1.098 ± 0.015 | 3.985 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
