
Ruminococcus sp. YE71
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Ruminococcus; unclassified Ruminococcus
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
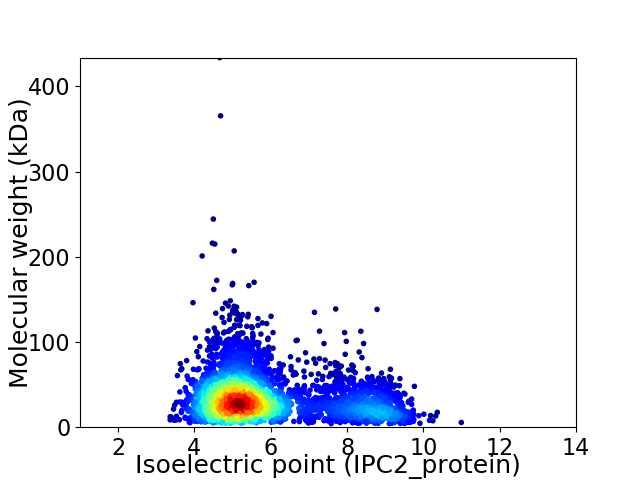
Virtual 2D-PAGE plot for 3572 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1K1MQ30|A0A1K1MQ30_9FIRM D-alanyl-D-alanine carboxypeptidase OS=Ruminococcus sp. YE71 OX=244362 GN=SAMN02910447_01086 PE=4 SV=1
MM1 pKa = 7.91KK2 pKa = 9.79KK3 pKa = 10.41RR4 pKa = 11.84KK5 pKa = 8.04MVCVLAAVLLLAGCGKK21 pKa = 8.17TEE23 pKa = 4.24KK24 pKa = 10.09EE25 pKa = 3.98AASASAEE32 pKa = 4.05DD33 pKa = 3.93TAQASADD40 pKa = 3.7AADD43 pKa = 4.14GDD45 pKa = 4.57VVSQEE50 pKa = 4.29SAAEE54 pKa = 3.91PAPEE58 pKa = 4.15NADD61 pKa = 3.44SSSVTDD67 pKa = 3.24STSDD71 pKa = 3.55YY72 pKa = 10.28KK73 pKa = 10.81IKK75 pKa = 10.59SHH77 pKa = 6.93GSRR80 pKa = 11.84EE81 pKa = 4.21YY82 pKa = 11.2NGAAANDD89 pKa = 4.02DD90 pKa = 3.81FADD93 pKa = 3.64ALFIGDD99 pKa = 3.86SRR101 pKa = 11.84TVGLMLNGDD110 pKa = 4.22KK111 pKa = 10.89PKK113 pKa = 10.87ADD115 pKa = 3.83YY116 pKa = 9.97FATAGLNVMTAQSTPVVPKK135 pKa = 10.16RR136 pKa = 11.84DD137 pKa = 3.75FEE139 pKa = 4.33GLIVQQAYY147 pKa = 8.78TDD149 pKa = 4.06YY150 pKa = 11.47GWGDD154 pKa = 3.76NAWDD158 pKa = 3.65DD159 pKa = 4.25TTWDD163 pKa = 3.56DD164 pKa = 4.08TTWDD168 pKa = 3.62DD169 pKa = 4.87GGWTGGDD176 pKa = 3.25STEE179 pKa = 4.13WDD181 pKa = 3.2WGYY184 pKa = 11.15SEE186 pKa = 5.3PDD188 pKa = 3.2SSEE191 pKa = 3.88EE192 pKa = 3.97TSSVDD197 pKa = 3.3EE198 pKa = 4.91DD199 pKa = 4.66LMTIPEE205 pKa = 4.26ALALKK210 pKa = 9.43QYY212 pKa = 10.88KK213 pKa = 10.14RR214 pKa = 11.84IFINFGVNEE223 pKa = 4.83LGWPYY228 pKa = 10.63PDD230 pKa = 3.73VFVEE234 pKa = 4.96RR235 pKa = 11.84YY236 pKa = 9.07KK237 pKa = 11.23LLILQVRR244 pKa = 11.84AAQPDD249 pKa = 3.48AQIYY253 pKa = 8.65IEE255 pKa = 4.43SMLPVSPLALSIDD268 pKa = 3.26KK269 pKa = 10.77CYY271 pKa = 8.73TTEE274 pKa = 4.09NVDD277 pKa = 3.75NFNEE281 pKa = 4.26NYY283 pKa = 9.19VRR285 pKa = 11.84RR286 pKa = 11.84AAQEE290 pKa = 4.2TNCTYY295 pKa = 11.34LDD297 pKa = 3.56VNQFFRR303 pKa = 11.84DD304 pKa = 3.3ADD306 pKa = 3.67GGLFADD312 pKa = 4.76ASTDD316 pKa = 4.07GIHH319 pKa = 7.41LNRR322 pKa = 11.84DD323 pKa = 3.3YY324 pKa = 11.57CMEE327 pKa = 3.8WMDD330 pKa = 3.39ILSYY334 pKa = 9.3YY335 pKa = 9.03TVGGGDD341 pKa = 3.14ISILSRR347 pKa = 11.84RR348 pKa = 11.84HH349 pKa = 4.4EE350 pKa = 4.42TEE352 pKa = 3.7SAPDD356 pKa = 3.88GSIEE360 pKa = 4.14PYY362 pKa = 10.53SDD364 pKa = 3.69GGDD367 pKa = 3.61GYY369 pKa = 11.33SEE371 pKa = 5.61DD372 pKa = 5.52GGTDD376 pKa = 2.91WTDD379 pKa = 3.16GGDD382 pKa = 3.81NGWGGDD388 pKa = 4.61GYY390 pKa = 11.28SDD392 pKa = 3.38WNDD395 pKa = 2.93EE396 pKa = 4.28GGYY399 pKa = 10.92GDD401 pKa = 4.14YY402 pKa = 11.14NYY404 pKa = 10.79GYY406 pKa = 10.97EE407 pKa = 4.98DD408 pKa = 3.39YY409 pKa = 10.45TYY411 pKa = 11.22DD412 pKa = 3.93NEE414 pKa = 3.87YY415 pKa = 10.96DD416 pKa = 3.44YY417 pKa = 11.11DD418 pKa = 3.7YY419 pKa = 11.48GYY421 pKa = 11.75
MM1 pKa = 7.91KK2 pKa = 9.79KK3 pKa = 10.41RR4 pKa = 11.84KK5 pKa = 8.04MVCVLAAVLLLAGCGKK21 pKa = 8.17TEE23 pKa = 4.24KK24 pKa = 10.09EE25 pKa = 3.98AASASAEE32 pKa = 4.05DD33 pKa = 3.93TAQASADD40 pKa = 3.7AADD43 pKa = 4.14GDD45 pKa = 4.57VVSQEE50 pKa = 4.29SAAEE54 pKa = 3.91PAPEE58 pKa = 4.15NADD61 pKa = 3.44SSSVTDD67 pKa = 3.24STSDD71 pKa = 3.55YY72 pKa = 10.28KK73 pKa = 10.81IKK75 pKa = 10.59SHH77 pKa = 6.93GSRR80 pKa = 11.84EE81 pKa = 4.21YY82 pKa = 11.2NGAAANDD89 pKa = 4.02DD90 pKa = 3.81FADD93 pKa = 3.64ALFIGDD99 pKa = 3.86SRR101 pKa = 11.84TVGLMLNGDD110 pKa = 4.22KK111 pKa = 10.89PKK113 pKa = 10.87ADD115 pKa = 3.83YY116 pKa = 9.97FATAGLNVMTAQSTPVVPKK135 pKa = 10.16RR136 pKa = 11.84DD137 pKa = 3.75FEE139 pKa = 4.33GLIVQQAYY147 pKa = 8.78TDD149 pKa = 4.06YY150 pKa = 11.47GWGDD154 pKa = 3.76NAWDD158 pKa = 3.65DD159 pKa = 4.25TTWDD163 pKa = 3.56DD164 pKa = 4.08TTWDD168 pKa = 3.62DD169 pKa = 4.87GGWTGGDD176 pKa = 3.25STEE179 pKa = 4.13WDD181 pKa = 3.2WGYY184 pKa = 11.15SEE186 pKa = 5.3PDD188 pKa = 3.2SSEE191 pKa = 3.88EE192 pKa = 3.97TSSVDD197 pKa = 3.3EE198 pKa = 4.91DD199 pKa = 4.66LMTIPEE205 pKa = 4.26ALALKK210 pKa = 9.43QYY212 pKa = 10.88KK213 pKa = 10.14RR214 pKa = 11.84IFINFGVNEE223 pKa = 4.83LGWPYY228 pKa = 10.63PDD230 pKa = 3.73VFVEE234 pKa = 4.96RR235 pKa = 11.84YY236 pKa = 9.07KK237 pKa = 11.23LLILQVRR244 pKa = 11.84AAQPDD249 pKa = 3.48AQIYY253 pKa = 8.65IEE255 pKa = 4.43SMLPVSPLALSIDD268 pKa = 3.26KK269 pKa = 10.77CYY271 pKa = 8.73TTEE274 pKa = 4.09NVDD277 pKa = 3.75NFNEE281 pKa = 4.26NYY283 pKa = 9.19VRR285 pKa = 11.84RR286 pKa = 11.84AAQEE290 pKa = 4.2TNCTYY295 pKa = 11.34LDD297 pKa = 3.56VNQFFRR303 pKa = 11.84DD304 pKa = 3.3ADD306 pKa = 3.67GGLFADD312 pKa = 4.76ASTDD316 pKa = 4.07GIHH319 pKa = 7.41LNRR322 pKa = 11.84DD323 pKa = 3.3YY324 pKa = 11.57CMEE327 pKa = 3.8WMDD330 pKa = 3.39ILSYY334 pKa = 9.3YY335 pKa = 9.03TVGGGDD341 pKa = 3.14ISILSRR347 pKa = 11.84RR348 pKa = 11.84HH349 pKa = 4.4EE350 pKa = 4.42TEE352 pKa = 3.7SAPDD356 pKa = 3.88GSIEE360 pKa = 4.14PYY362 pKa = 10.53SDD364 pKa = 3.69GGDD367 pKa = 3.61GYY369 pKa = 11.33SEE371 pKa = 5.61DD372 pKa = 5.52GGTDD376 pKa = 2.91WTDD379 pKa = 3.16GGDD382 pKa = 3.81NGWGGDD388 pKa = 4.61GYY390 pKa = 11.28SDD392 pKa = 3.38WNDD395 pKa = 2.93EE396 pKa = 4.28GGYY399 pKa = 10.92GDD401 pKa = 4.14YY402 pKa = 11.14NYY404 pKa = 10.79GYY406 pKa = 10.97EE407 pKa = 4.98DD408 pKa = 3.39YY409 pKa = 10.45TYY411 pKa = 11.22DD412 pKa = 3.93NEE414 pKa = 3.87YY415 pKa = 10.96DD416 pKa = 3.44YY417 pKa = 11.11DD418 pKa = 3.7YY419 pKa = 11.48GYY421 pKa = 11.75
Molecular weight: 46.35 kDa
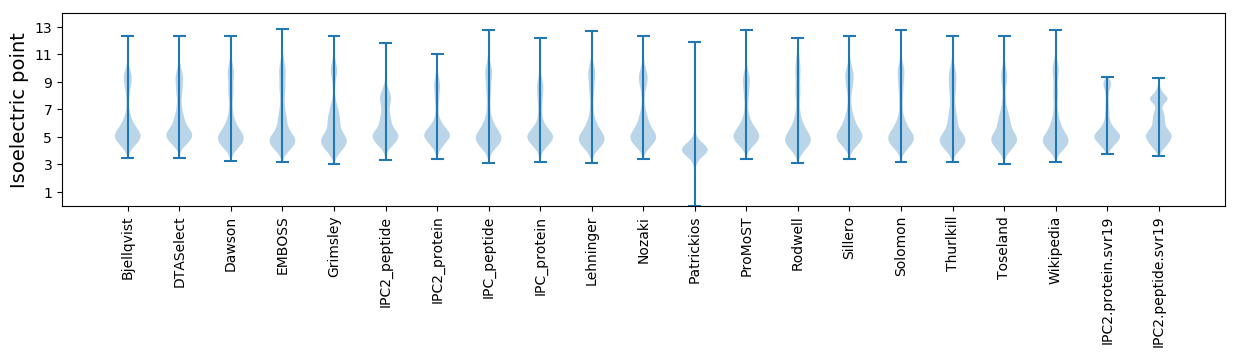
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1K1LMB9|A0A1K1LMB9_9FIRM Type IV pilus assembly protein PilB OS=Ruminococcus sp. YE71 OX=244362 GN=SAMN02910447_00223 PE=3 SV=1
MM1 pKa = 7.62TDD3 pKa = 2.38IKK5 pKa = 10.41TRR7 pKa = 11.84LQYY10 pKa = 10.51SAAVAAVILLGLVSRR25 pKa = 11.84RR26 pKa = 11.84IPVIPPCVGDD36 pKa = 4.1ALWAVVVFFCWRR48 pKa = 11.84IVLVKK53 pKa = 9.85KK54 pKa = 10.2HH55 pKa = 6.17PRR57 pKa = 11.84TAAAAALVTSFAVEE71 pKa = 4.31FSQLIRR77 pKa = 11.84WDD79 pKa = 2.97WLVRR83 pKa = 11.84VRR85 pKa = 11.84ATTIGHH91 pKa = 7.32LLLGQGFVWSDD102 pKa = 2.99LAAYY106 pKa = 9.76AVGIGAALLCVTAVGRR122 pKa = 11.84SKK124 pKa = 11.34
MM1 pKa = 7.62TDD3 pKa = 2.38IKK5 pKa = 10.41TRR7 pKa = 11.84LQYY10 pKa = 10.51SAAVAAVILLGLVSRR25 pKa = 11.84RR26 pKa = 11.84IPVIPPCVGDD36 pKa = 4.1ALWAVVVFFCWRR48 pKa = 11.84IVLVKK53 pKa = 9.85KK54 pKa = 10.2HH55 pKa = 6.17PRR57 pKa = 11.84TAAAAALVTSFAVEE71 pKa = 4.31FSQLIRR77 pKa = 11.84WDD79 pKa = 2.97WLVRR83 pKa = 11.84VRR85 pKa = 11.84ATTIGHH91 pKa = 7.32LLLGQGFVWSDD102 pKa = 2.99LAAYY106 pKa = 9.76AVGIGAALLCVTAVGRR122 pKa = 11.84SKK124 pKa = 11.34
Molecular weight: 13.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1169459 |
39 |
3788 |
327.4 |
36.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.075 ± 0.045 | 1.689 ± 0.017 |
6.419 ± 0.035 | 7.156 ± 0.04 |
4.218 ± 0.029 | 7.194 ± 0.041 |
1.625 ± 0.015 | 6.785 ± 0.037 |
6.382 ± 0.034 | 8.41 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.805 ± 0.02 | 4.236 ± 0.037 |
3.348 ± 0.021 | 2.609 ± 0.023 |
4.659 ± 0.036 | 6.639 ± 0.042 |
6.0 ± 0.042 | 6.712 ± 0.029 |
0.902 ± 0.015 | 4.134 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
