
Wenling tombus-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.73
Get precalculated fractions of proteins
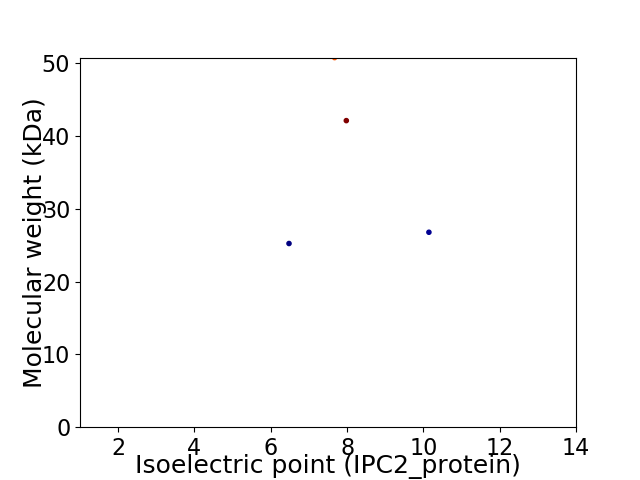
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

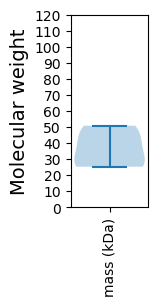
Protein with the lowest isoelectric point:
>tr|A0A1L3KH40|A0A1L3KH40_9VIRU Uncharacterized protein OS=Wenling tombus-like virus 2 OX=1923544 PE=4 SV=1
MM1 pKa = 6.77YY2 pKa = 10.8AEE4 pKa = 4.73IEE6 pKa = 4.34SAAWVYY12 pKa = 11.41NKK14 pKa = 10.85AKK16 pKa = 9.94FVWKK20 pKa = 9.25MYY22 pKa = 10.12PVLRR26 pKa = 11.84DD27 pKa = 3.39DD28 pKa = 3.7TFILYY33 pKa = 10.3RR34 pKa = 11.84SVEE37 pKa = 4.09LPKK40 pKa = 10.78LKK42 pKa = 10.63SFQTYY47 pKa = 10.64CNITGTISSSQYY59 pKa = 9.85YY60 pKa = 10.49GFDD63 pKa = 3.07EE64 pKa = 4.3VTLNLRR70 pKa = 11.84VEE72 pKa = 4.38RR73 pKa = 11.84SKK75 pKa = 10.9DD76 pKa = 2.98WKK78 pKa = 10.78VIEE81 pKa = 4.25EE82 pKa = 4.37SMVTLDD88 pKa = 3.26SHH90 pKa = 6.49TPGNLFPTDD99 pKa = 3.12NRR101 pKa = 11.84SAVQLLYY108 pKa = 10.41TGTFEE113 pKa = 5.01VPPDD117 pKa = 3.36HH118 pKa = 7.26HH119 pKa = 6.71ITTSSILAVIARR131 pKa = 11.84YY132 pKa = 8.51PVVATTTLSLAGRR145 pKa = 11.84SAWDD149 pKa = 3.28ILRR152 pKa = 11.84MYY154 pKa = 9.91FKK156 pKa = 11.14VSFHH160 pKa = 6.82SSLVFNIKK168 pKa = 10.46GEE170 pKa = 4.15EE171 pKa = 3.92NIKK174 pKa = 10.06IVRR177 pKa = 11.84SASFPMLSHH186 pKa = 7.12RR187 pKa = 11.84SLASSSSWQSLPEE200 pKa = 4.07LSSGRR205 pKa = 11.84SSAQLSSVSLNSEE218 pKa = 3.5EE219 pKa = 4.31WIAII223 pKa = 3.82
MM1 pKa = 6.77YY2 pKa = 10.8AEE4 pKa = 4.73IEE6 pKa = 4.34SAAWVYY12 pKa = 11.41NKK14 pKa = 10.85AKK16 pKa = 9.94FVWKK20 pKa = 9.25MYY22 pKa = 10.12PVLRR26 pKa = 11.84DD27 pKa = 3.39DD28 pKa = 3.7TFILYY33 pKa = 10.3RR34 pKa = 11.84SVEE37 pKa = 4.09LPKK40 pKa = 10.78LKK42 pKa = 10.63SFQTYY47 pKa = 10.64CNITGTISSSQYY59 pKa = 9.85YY60 pKa = 10.49GFDD63 pKa = 3.07EE64 pKa = 4.3VTLNLRR70 pKa = 11.84VEE72 pKa = 4.38RR73 pKa = 11.84SKK75 pKa = 10.9DD76 pKa = 2.98WKK78 pKa = 10.78VIEE81 pKa = 4.25EE82 pKa = 4.37SMVTLDD88 pKa = 3.26SHH90 pKa = 6.49TPGNLFPTDD99 pKa = 3.12NRR101 pKa = 11.84SAVQLLYY108 pKa = 10.41TGTFEE113 pKa = 5.01VPPDD117 pKa = 3.36HH118 pKa = 7.26HH119 pKa = 6.71ITTSSILAVIARR131 pKa = 11.84YY132 pKa = 8.51PVVATTTLSLAGRR145 pKa = 11.84SAWDD149 pKa = 3.28ILRR152 pKa = 11.84MYY154 pKa = 9.91FKK156 pKa = 11.14VSFHH160 pKa = 6.82SSLVFNIKK168 pKa = 10.46GEE170 pKa = 4.15EE171 pKa = 3.92NIKK174 pKa = 10.06IVRR177 pKa = 11.84SASFPMLSHH186 pKa = 7.12RR187 pKa = 11.84SLASSSSWQSLPEE200 pKa = 4.07LSSGRR205 pKa = 11.84SSAQLSSVSLNSEE218 pKa = 3.5EE219 pKa = 4.31WIAII223 pKa = 3.82
Molecular weight: 25.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGP2|A0A1L3KGP2_9VIRU Uncharacterized protein OS=Wenling tombus-like virus 2 OX=1923544 PE=4 SV=1
MM1 pKa = 7.62EE2 pKa = 4.88IVPRR6 pKa = 11.84GRR8 pKa = 11.84RR9 pKa = 11.84APPLGTVQAALRR21 pKa = 11.84GLNLARR27 pKa = 11.84QAYY30 pKa = 9.32NWMNPAPQQPQNPPGTGRR48 pKa = 11.84RR49 pKa = 11.84SRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84GGRR56 pKa = 11.84GRR58 pKa = 11.84GVSTPVQPIPMPIYY72 pKa = 9.23PAFPRR77 pKa = 11.84GRR79 pKa = 11.84GRR81 pKa = 11.84GRR83 pKa = 11.84GRR85 pKa = 11.84GRR87 pKa = 11.84VLTTNPGRR95 pKa = 11.84SVTITQSEE103 pKa = 4.55VWGDD107 pKa = 3.62VVKK110 pKa = 10.97GPNVFTFQPSAIGMPQLKK128 pKa = 9.66HH129 pKa = 5.98LAAGYY134 pKa = 7.93TRR136 pKa = 11.84YY137 pKa = 10.32KK138 pKa = 9.82VIRR141 pKa = 11.84VTLEE145 pKa = 3.38YY146 pKa = 9.98EE147 pKa = 4.05PAVGRR152 pKa = 11.84QRR154 pKa = 11.84DD155 pKa = 4.15GVIHH159 pKa = 6.04WGIRR163 pKa = 11.84PGVVAKK169 pKa = 9.41ATATPEE175 pKa = 4.14VCKK178 pKa = 10.09AAYY181 pKa = 8.2PRR183 pKa = 11.84RR184 pKa = 11.84SHH186 pKa = 6.16PVYY189 pKa = 10.78VRR191 pKa = 11.84TTMTIGHH198 pKa = 7.26DD199 pKa = 4.71LIMMQPWLGTGEE211 pKa = 4.13TAFTLVANSPGDD223 pKa = 3.47TTQPGFFEE231 pKa = 4.36CTYY234 pKa = 10.77HH235 pKa = 7.03VVLDD239 pKa = 4.18LPKK242 pKa = 10.49PPP244 pKa = 4.46
MM1 pKa = 7.62EE2 pKa = 4.88IVPRR6 pKa = 11.84GRR8 pKa = 11.84RR9 pKa = 11.84APPLGTVQAALRR21 pKa = 11.84GLNLARR27 pKa = 11.84QAYY30 pKa = 9.32NWMNPAPQQPQNPPGTGRR48 pKa = 11.84RR49 pKa = 11.84SRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84GGRR56 pKa = 11.84GRR58 pKa = 11.84GVSTPVQPIPMPIYY72 pKa = 9.23PAFPRR77 pKa = 11.84GRR79 pKa = 11.84GRR81 pKa = 11.84GRR83 pKa = 11.84GRR85 pKa = 11.84GRR87 pKa = 11.84VLTTNPGRR95 pKa = 11.84SVTITQSEE103 pKa = 4.55VWGDD107 pKa = 3.62VVKK110 pKa = 10.97GPNVFTFQPSAIGMPQLKK128 pKa = 9.66HH129 pKa = 5.98LAAGYY134 pKa = 7.93TRR136 pKa = 11.84YY137 pKa = 10.32KK138 pKa = 9.82VIRR141 pKa = 11.84VTLEE145 pKa = 3.38YY146 pKa = 9.98EE147 pKa = 4.05PAVGRR152 pKa = 11.84QRR154 pKa = 11.84DD155 pKa = 4.15GVIHH159 pKa = 6.04WGIRR163 pKa = 11.84PGVVAKK169 pKa = 9.41ATATPEE175 pKa = 4.14VCKK178 pKa = 10.09AAYY181 pKa = 8.2PRR183 pKa = 11.84RR184 pKa = 11.84SHH186 pKa = 6.16PVYY189 pKa = 10.78VRR191 pKa = 11.84TTMTIGHH198 pKa = 7.26DD199 pKa = 4.71LIMMQPWLGTGEE211 pKa = 4.13TAFTLVANSPGDD223 pKa = 3.47TTQPGFFEE231 pKa = 4.36CTYY234 pKa = 10.77HH235 pKa = 7.03VVLDD239 pKa = 4.18LPKK242 pKa = 10.49PPP244 pKa = 4.46
Molecular weight: 26.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1276 |
223 |
443 |
319.0 |
36.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.661 ± 0.214 | 1.019 ± 0.166 |
4.624 ± 0.834 | 5.721 ± 1.026 |
3.448 ± 0.273 | 5.721 ± 1.454 |
2.665 ± 0.223 | 5.486 ± 0.38 |
6.27 ± 1.366 | 7.915 ± 0.775 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.056 ± 0.211 | 3.213 ± 0.101 |
6.505 ± 1.373 | 4.389 ± 0.775 |
6.426 ± 1.188 | 7.602 ± 1.6 |
6.113 ± 0.762 | 7.132 ± 0.751 |
1.803 ± 0.189 | 4.232 ± 0.278 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
